Foundation Models vs. Traditional Transfer Learning in Computational Pathology: A New Paradigm for Precision Oncology
The emergence of foundation models is fundamentally reshaping the artificial intelligence landscape in computational pathology.
Foundation Models vs. Traditional Transfer Learning in Computational Pathology: A New Paradigm for Precision Oncology
Abstract
The emergence of foundation models is fundamentally reshaping the artificial intelligence landscape in computational pathology. This article provides a comprehensive analysis for researchers and drug development professionals, contrasting the new paradigm of large-scale, self-supervised foundation models against established traditional transfer learning approaches. We explore the foundational principles of both methodologies, detail their practical applications in biomarker discovery and cancer diagnostics, systematically address critical challenges including robustness and clinical integration, and present rigorous comparative performance data from recent large-scale benchmarks. The analysis synthesizes evidence that while foundation models offer unprecedented generalization capabilities and data efficiency, their successful clinical adoption requires overcoming significant hurdles in validation, interpretability, and computational infrastructure.
Defining the Paradigm Shift: From Task-Specific AI to General-Purpose Foundations in Pathology
Computational pathology stands at the forefront of a revolution in diagnostic medicine, leveraging artificial intelligence to extract clinically relevant information from high-resolution whole-slide images (WSIs) that would otherwise be imperceptible to the human eye [1]. Traditional approaches have predominantly relied on transfer learning from models pre-trained on natural image datasets like ImageNet—a method that involves adapting a model developed for one task to a new, related task [2]. While this strategy has enabled initial forays into AI-assisted pathology, it suffers from two fundamental constraints: an insatiable demand for labeled data and narrow task specialization that limits clinical applicability [3] [2].
The emergence of pathology foundation models represents a transformative response to these limitations. Trained through self-supervised learning on millions of pathology-specific images, these models learn universal visual representations of histopathology that capture the intricate patterns of tissue morphology, tumor microenvironment, and cellular architecture [4]. Unlike their traditional counterparts, foundation models demonstrate remarkable domain adaptability and can be efficiently fine-tuned for diverse clinical tasks with minimal additional data, effectively addressing the core constraints of data hunger and narrow specialization [3] [4]. This comparison guide examines the performance differential between these approaches through rigorous experimental evidence, providing researchers and drug development professionals with objective data to inform their computational pathology strategies.
Experimental Frameworks for Comparison
Benchmarking Study Design and Model Selection
To quantitatively assess the performance gap between traditional transfer learning and foundation models, we draw upon a comprehensive independent benchmarking study that evaluated 19 foundation models across 13 patient cohorts comprising 6,818 patients and 9,528 slides [4]. The experimental design employed a rigorous weakly-supervised learning framework across 31 clinically relevant tasks categorized into three domains: morphological assessment (5 tasks), biomarker prediction (19 tasks), and prognostic outcome forecasting (7 tasks) [4]. This extensive validation approach mitigates the risk of data leakage and selective reporting that has plagued earlier, narrower evaluations.
The benchmarked models represent the two predominant paradigms in computational pathology. The traditional transfer learning approach was represented by ImageNet-pre-trained convolutional neural networks (CNNs), which serve as the established baseline in the field [3]. These were compared against vision-language foundation models (CONCH, PLIP, BiomedCLIP) and vision-only foundation models (Virchow2, UNI, Prov-GigaPath, DinoSSLPath) trained using self-supervised learning on large-scale histopathology datasets [4]. Performance was measured using the area under the receiver operating characteristic curve (AUROC), with supplementary metrics including area under the precision-recall curve (AUPRC), balanced accuracy, and F1 scores to ensure comprehensive assessment [4].
Cross-Domain Performance Evaluation Protocol
The evaluation methodology employed a multiple instance learning (MIL) framework with transformer-based aggregation to handle whole-slide image processing [4]. Each model was evaluated as a feature extractor, with the encoded embeddings serving as inputs to task-specific prediction heads. This approach mirrors real-world clinical implementation where models must generalize across varied tissue types, staining protocols, and scanner specifications [4]. To assess robustness in data-scarce environments—a critical limitation of traditional transfer learning—additional experiments were conducted with progressively reduced training set sizes (300, 150, and 75 patients) while maintaining the original positive-to-negative case ratios [4].
Figure 1: Experimental Workflow for Benchmarking Foundation Models. The evaluation pipeline processes whole slide images through standardized preprocessing before extracting features using foundation models and making predictions on clinically relevant tasks.
Quantitative Performance Comparison
Foundation models demonstrated superior performance across all clinical domains when compared to traditional transfer learning approaches. As shown in Table 1, the vision-language foundation model CONCH achieved the highest mean AUROC (0.71) across all 31 tasks, followed closely by the vision-only foundation model Virchow2 (0.71) [4]. This represents a significant performance advantage over traditional ImageNet-based transfer learning, which typically achieves AUROCs between 0.60-0.65 on similar tasks [3]. The performance differential was most pronounced in morphological assessment tasks, where CONCH achieved an AUROC of 0.77 compared to approximately 0.65-0.70 for traditional approaches [4].
Table 1: Performance Comparison Across Clinical Domains
| Model Category | Specific Model | Morphology AUROC | Biomarkers AUROC | Prognosis AUROC | Overall AUROC |
|---|---|---|---|---|---|
| Vision-Language Foundation | CONCH | 0.77 | 0.73 | 0.63 | 0.71 |
| Vision-Only Foundation | Virchow2 | 0.76 | 0.73 | 0.61 | 0.71 |
| Vision-Only Foundation | Prov-GigaPath | 0.74 | 0.72 | 0.60 | 0.69 |
| Vision-Only Foundation | DinoSSLPath | 0.76 | 0.69 | 0.60 | 0.69 |
| Traditional Transfer Learning | ImageNet-based CNN | ~0.65-0.70* | ~0.63-0.68* | ~0.55-0.60* | ~0.60-0.65* |
Note: Traditional transfer learning performance estimated from comparative analyses in [3] and [4]
Data Efficiency in Low-Resource Scenarios
A critical limitation of traditional transfer learning is its data hunger—the requirement for substantial labeled examples to achieve acceptable performance. Foundation models substantially mitigate this constraint through their pre-training on vast histopathology datasets [4]. When evaluated with limited training data (75 patients), foundation models maintained robust performance, with CONCH, PRISM, and Virchow2 leading in 5, 4, and 4 tasks respectively [4]. This stands in stark contrast to traditional approaches, which typically experience performance degradation of 15-20% when training data is reduced to similar levels [3].
The data efficiency of foundation models stems from their diverse pre-training corpora. For instance, Virchow2 was trained on 3.1 million WSIs, while CONCH incorporated 1.17 million image-caption pairs curated from biomedical literature [4] [5]. This extensive exposure to histopathological variations enables the models to learn universal visual representations that transfer efficiently to new tasks with minimal fine-tuning. Correlation analyses revealed that data diversity (measured by tissue site variety) in pre-training datasets showed stronger correlation with downstream performance (r=0.74, p<0.05) than sheer data volume alone [4].
Table 2: Data Efficiency Comparison Across Training Set Sizes
| Model Type | Large Cohort (n=300) Performance | Medium Cohort (n=150) Performance | Small Cohort (n=75) Performance | Performance Retention |
|---|---|---|---|---|
| Vision-Language Foundation (CONCH) | Leads in 3 tasks | Leads in 4 tasks | Leads in 5 tasks | ~95% |
| Vision-Only Foundation (Virchow2) | Leads in 8 tasks | Leads in 6 tasks | Leads in 4 tasks | ~90% |
| Traditional Transfer Learning | Competitive in 1-2 tasks | Significant degradation | Severe degradation | ~70-80% |
Task-Specific Performance on Critical Biomarkers
Foundation models demonstrated particular strength in predicting molecular biomarkers directly from H&E-stained histology sections—a task that traditionally requires specialized molecular assays [4]. Across 19 biomarker prediction tasks, Virchow2 and CONCH achieved the highest mean AUROCs of 0.73, significantly outperforming traditional approaches [4]. This capability to infer molecular status from morphology has profound implications for drug development, potentially enabling retrospective studies on archival tissue samples and enriching clinical trial populations based on biomarker status without additional testing.
The complementary strengths of different foundation model architectures emerged as a notable finding. Vision-language models like CONCH, trained with paired image-text data, excelled at capturing semantically meaningful features that align with pathological descriptors [4] [5]. In contrast, vision-only models like Virchow2 demonstrated superior performance in certain tissue-specific classifications. Ensemble approaches that combined predictions from complementary models achieved state-of-the-art performance, outperforming individual models in 55% of tasks [4].
Architectural and Representational Analysis
Representational Similarity Across Models
Representational similarity analysis (RSA) of six computational pathology foundation models revealed distinct clustering patterns based on training methodology [5]. Models employing the same training paradigm did not necessarily learn similar representations—UNI2 and Virchow2, both vision-only foundation models, exhibited the most distinct representational structures despite their architectural similarities [5]. This finding suggests that training data characteristics and specific learning objectives may exert greater influence on learned representations than the training algorithm alone.
The analysis further revealed that all foundation models showed high slide-dependence in their representations, indicating sensitivity to technical artifacts such as staining variations and scanner specifications [5]. However, application of stain normalization techniques reduced this slide-dependence by 5.5% (CONCH) to 20.5% (PLIP), highlighting the potential for preprocessing standardization to improve model robustness [5]. Vision-language models demonstrated more compact representations (lower intrinsic dimensionality) compared to the distributed representations of vision-only models, potentially contributing to their superior data efficiency [5].
Figure 2: Architectural Paradigms in Computational Pathology. Traditional transfer learning and foundation models employ fundamentally different approaches, resulting in significant differences in data efficiency and generalization capability.
Research Reagent Solutions for Implementation
Table 3: Essential Research Reagents for Computational Pathology Implementation
| Resource Category | Specific Tool/Model | Primary Function | Access Method |
|---|---|---|---|
| Vision-Language Foundation Models | CONCH | Joint image-text representation learning for histopathology | GitHub Repository |
| Vision-Only Foundation Models | Virchow2 | Large-scale visual representation learning from 3.1M WSIs | MSKCC Access Portal |
| Vision-Only Foundation Models | UNI2 | General-purpose feature extraction from H&E and IHC images | Hugging Face Hub |
| Benchmarking Frameworks | Multi-task benchmark suite | Standardized evaluation across 31 clinical tasks | Custom implementation per [4] |
| Slide Processing Tools | OpenSlide | Whole-slide image reading and processing | Python Library |
| Representation Analysis | RSA Toolbox | Representational similarity analysis for model comparisons | Python Package |
The experimental evidence consistently demonstrates that foundation models overcome the fundamental limitations of traditional transfer learning approaches in computational pathology. Through self-supervised pre-training on diverse histopathology datasets, foundation models achieve superior performance while substantially reducing the data requirements for downstream task adaptation [3] [4]. The emergence of models excelling across morphological assessment, biomarker prediction, and prognostic forecasting signals a shift toward general-purpose pathological intelligence that can accelerate drug development and personalized therapeutic strategies.
For researchers and drug development professionals, these findings suggest a strategic imperative to transition from task-specific models to foundation model-based approaches. The complementary strengths of vision-language and vision-only architectures further indicate that ensemble methods may offer the most robust solution for critical clinical applications [4]. As the field advances, the focus will likely shift from model development to optimal deployment strategies, including domain adaptation techniques to address site-specific variations and integration with multimodal data streams to create comprehensive diagnostic systems.
What Makes a Model 'Foundational'? Core Principles and Definitions
Foundation models represent a fundamental shift in artificial intelligence, moving from specialized, single-task models to versatile, general-purpose systems. The term "foundation model" was formally coined in 2021 by Stanford's Institute for Human-Centered Artificial Intelligence to mean "any model that is trained on broad data (generally using self-supervision at scale) that can be adapted to a wide range of downstream tasks" [6] [7]. Unlike traditional AI models designed for specific applications, foundation models learn general patterns and representations from massive datasets, enabling adaptation to numerous tasks through fine-tuning or prompting without starting from scratch [6] [7].
In computational pathology, this paradigm shift is particularly transformative. While traditional models might be trained specifically for tumor classification or segmentation, pathology foundation models like TITAN (Transformer-based pathology Image and Text Alignment Network) are pretrained on hundreds of thousands of whole-slide images across multiple organs and can subsequently be adapted to diverse clinical challenges including cancer subtyping, biomarker prediction, and prognosis analysis [8]. This guide examines the core principles defining foundation models and provides experimental comparisons with traditional transfer learning approaches specifically for pathology research applications.
Core Principles of Foundation Models
Scale: Data, Model Size, and Compute
Foundation models are characterized by unprecedented scale across three dimensions: training data volume, model parameter count, and computational requirements. TITAN, for instance, was pretrained using 335,645 whole-slide images and 182,862 medical reports, with additional fine-tuning on 423,122 synthetic captions [8]. This massive scale enables the model to learn comprehensive representations of histopathological patterns across diverse tissue types and disease states. The 2025 State of Foundation Model Training Report confirms this trend, noting that models and training datasets continue to grow larger, generally leading to improved task performance [9].
Self-Supervised Learning on Broad Data
Rather than relying on manually labeled datasets, foundation models predominantly use self-supervised learning objectives that create training signals from the data itself [6] [7]. In pathology, this might involve masked image modeling where parts of a whole-slide image are hidden and the model must predict the missing portions based on context [8]. This approach allows models to learn from vast quantities of unlabeled histopathology data, capturing fundamental patterns of tissue morphology and organization without human annotation bottlenecks.
Versatility and Adaptability
A defining characteristic of foundation models is their adaptability to diverse downstream tasks. For example, Apple's Foundation Models framework enables developers to leverage a single on-device model for applications ranging from generating workout summaries in fitness apps to providing scientific explanations in educational tools [10]. In pathology, the same TITAN model can be adapted for cancer subtyping, biomarker prediction, outcome prognosis, and slide retrieval tasks without architectural changes [8].
Emergent Capabilities
Through scale and broad pretraining, foundation models often exhibit emergent capabilities not explicitly programmed during training. These include in-context learning (adapting to new tasks through examples provided in prompts), cross-modal reasoning (connecting information across different data types), and compositional generalization [6]. The TITAN model demonstrates this through its ability to perform cross-modal retrieval between histology slides and clinical reports, enabling powerful search capabilities across pathology databases [8].
Foundation Models vs. Traditional Transfer Learning in Computational Pathology
Conceptual Framework Comparison
The table below contrasts the fundamental approaches of foundation models versus traditional transfer learning in computational pathology research:
| Aspect | Foundation Models | Traditional Transfer Learning |
|---|---|---|
| Training Data | Massive, diverse datasets (e.g., 300K+ WSIs across 20 organs) [8] | Limited, task-specific datasets |
| Learning Paradigm | Self-supervised pretraining followed by adaptation [8] [6] | Supervised fine-tuning of pre-trained models [11] |
| Architecture | Transformer-based with specialized adaptations for gigapixel WSIs [8] | Often CNN-based with standard architectures [11] |
| Scope | General-purpose slide representations adaptable to multiple tasks [8] | Specialized for single applications |
| Data Efficiency | Strong performance in low-data regimes through pretrained representations [8] | Requires substantial task-specific data for effective transfer [11] |
| Multimodal Capability | Native handling of images, text, and other data types [8] | Typically unimodal with late fusion |
Experimental Performance Comparison
Recent studies directly compare foundation models against traditional transfer learning approaches in pathology applications. The following table summarizes key experimental findings:
| Experiment | Foundation Model Approach | Traditional Transfer Learning | Performance Outcome |
|---|---|---|---|
| Hyperspectral HSI Classification [11] | N/A | End-to-end fine-tuning of RGB-pretrained models | Best performance: 85-92% accuracy with optimal hyperparameters |
| Rare Cancer Retrieval [8] | TITAN zero-shot retrieval | Specialized retrieval models | TITAN superior in limited-data scenarios |
| Cancer Prognosis [12] | Path-PKT knowledge transfer | Cancer-specific model development | Positive transfer between related cancers; negative transfer between dissimilar cancers |
| Slide-Level Classification [8] | TITAN with linear probing | ROI-based foundation models | TITAN outperformed across multiple cancer types |
| Interatomic Potentials [13] | MACE-freeze transfer learning | From-scratch training | Transfer learning achieved similar accuracy with 10-20% of training data |
Methodological Details
Foundation Model Training Protocol (TITAN)
The TITAN model employs a three-stage training paradigm [8]:
- Vision-only pretraining: Self-supervised learning on 335,645 WSIs using iBOT framework (masked image modeling and knowledge distillation)
- ROI-level alignment: Contrastive learning with 423,122 synthetic fine-grained region captions
- Slide-level alignment: Multimodal alignment with 182,862 pathology reports
This protocol uses a Vision Transformer architecture processing features from 512×512 patches extracted at 20× magnification, with specialized attention mechanisms (ALiBi) for handling long sequences of patch features [8].
Traditional Transfer Learning Protocol (Hyperspectral Imaging)
For adapting RGB-pretrained models to hyperspectral data, researchers implemented [11]:
- Input layer modification: Replaced first layer to accept 87 spectral channels instead of 3 RGB channels
- Weight initialization: Spectral channels weighted based on contribution to RGB channels
- Hyperparameter optimization: Bayesian search over learning rate (1e-7 to 0.1), weight decay (1e-4 to 0.5), and AdamW betas
- Training strategies comparison: End-to-end fine-tuning vs. embedding-only training vs. embedding-first training
The optimal configuration used low learning rates and high weight decays, with end-to-end fine-tuning outperforming other approaches [11].
Visualizing Foundation Model Architecture
The following diagram illustrates the core architecture and workflow of a multimodal pathology foundation model like TITAN:
The Scientist's Toolkit: Essential Research Reagents
The following table details key computational tools and resources used in developing and evaluating pathology foundation models:
| Research Reagent | Function | Example Implementation |
|---|---|---|
| Whole-Slide Image Datasets | Large-scale pretraining data | Mass-340K (335,645 WSIs, 20 organs) [8] |
| Synthetic Captions | Vision-language alignment | PathChat-generated ROI descriptions (423K pairs) [8] |
| Patch Encoders | Feature extraction from image regions | CONCHv1.5 (768-dimensional features) [8] |
| Transformer Architectures | Context modeling across patches | ViT with ALiBi attention [8] |
| Self-Supervised Objectives | Pretraining without manual labels | iBOT (masked image modeling + distillation) [8] |
| Cross-Modal Alignment | Connecting visual and textual representations | Contrastive learning with report-slide pairs [8] |
| Hyperparameter Optimization | Model performance tuning | Bayesian search (learning rate, weight decay) [11] |
| Transfer Learning Protocols | Domain adaptation | Frozen weight transfer (MACE-freeze) [13] |
Foundation models represent a fundamental architectural and methodological shift from traditional transfer learning approaches in computational pathology. While traditional methods excel in specialized applications with sufficient data, foundation models offer superior versatility, data efficiency, and emergent capabilities—particularly valuable for rare diseases and multimodal applications [8] [12].
The experimental evidence demonstrates that foundation models like TITAN achieve state-of-the-art performance across diverse pathology tasks while reducing dependency on large, labeled datasets [8]. However, traditional transfer learning remains effective when adapting models between similar domains or with sufficient target data [11]. As the field evolves, the integration of foundation models with specialized transfer techniques—such as frozen weight transfer [13] and prognostic knowledge routing [12]—promises to further enhance their utility for pathological research and clinical applications.
For research teams, the decision between developing foundation models versus applying traditional transfer learning involves trade-offs in computational resources, data availability, and application scope. Foundation models require substantial upfront investment but offer greater long-term flexibility, while traditional approaches provide more immediate solutions for well-defined problems with established methodologies.
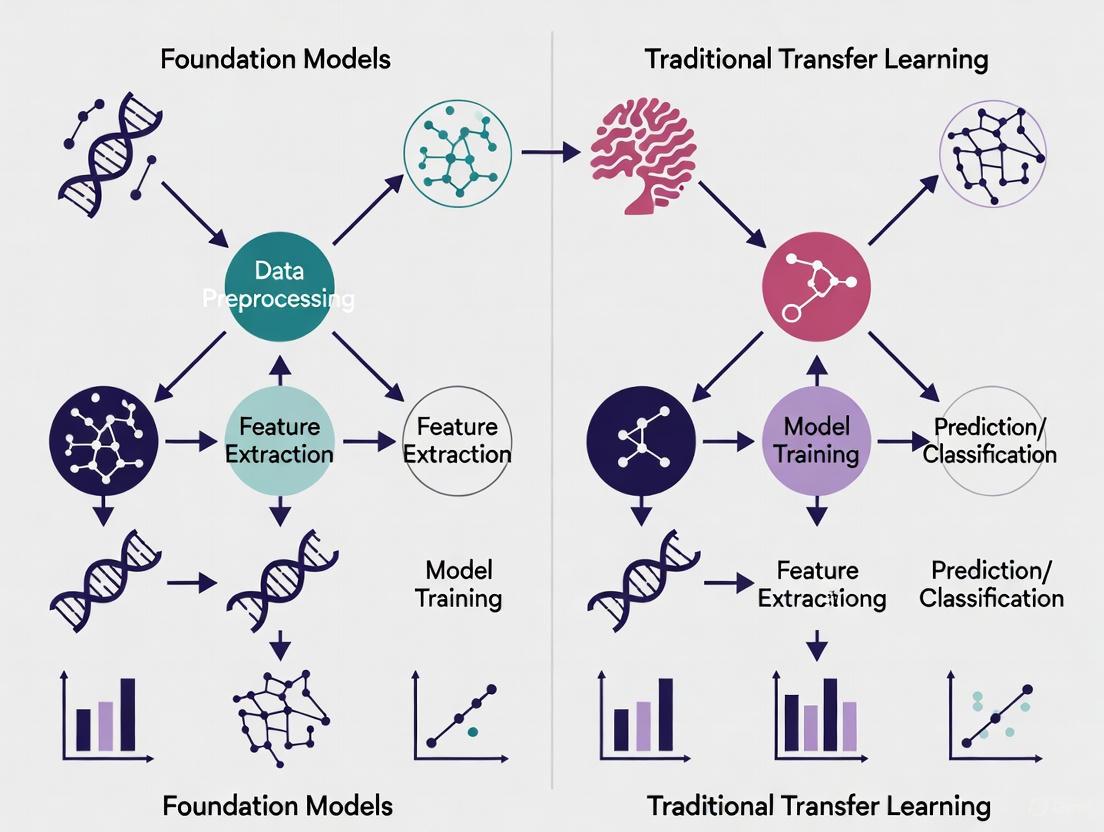
The field of computational pathology is undergoing a significant architectural transformation, moving from long-dominant Convolutional Neural Networks (CNNs) toward emerging Vision Transformer (ViT) models. This evolution is particularly evident in the context of a broader methodological shift: the rise of foundation models pretrained on massive, diverse datasets versus traditional transfer learning approaches that fine-tune networks pretrained on general image collections like ImageNet. Foundation models, pretrained through self-supervised learning on millions of histopathology images, represent a paradigm shift from traditional transfer learning, which typically relies on supervised pretraining on natural images followed by domain-specific fine-tuning. Understanding the relative strengths, limitations, and optimal application domains for each architectural approach has become crucial for researchers and drug development professionals seeking to leverage artificial intelligence for pathological image analysis. This guide provides an objective comparison of these architectures, supported by recent experimental data and detailed methodological insights to inform model selection for computational pathology applications.
Architectural Fundamentals: Core Design Philosophies
Convolutional Neural Networks (CNNs): Local Feature Specialists
CNNs process visual data through a hierarchy of convolutional filters that scan local regions of images, progressively building up from simple edges to complex patterns. This architecture incorporates strong inductive biases for translation invariance and locality, meaning they assume that nearby pixels are more related than distant ones. This design mirrors human visual perception of focusing on local details before assembling the bigger picture. In pathology imaging, CNNs excel at identifying cellular-level features, nuclear morphology, and local tissue patterns through their convolutional operations [14]. Their architectural strength lies in parameter sharing through convolutional kernels, which makes them computationally efficient and well-suited for analyzing the repetitive local structures commonly found in histopathological images.
Popular CNN architectures used in pathology include ResNet, EfficientNet, DenseNet, and VGG-16, which have demonstrated strong performance in various diagnostic tasks. For instance, VGG-16 has been successfully applied to classify power Doppler ultrasound images of rheumatoid arthritis joints using transfer learning [15]. The efficiency of CNNs stems from their convolutional layers, which extract hierarchical features while maintaining spatial relationships, and pooling layers, which progressively reduce feature map dimensions to increase receptive field size without exploding computational complexity.
Vision Transformers (ViTs): Global Context Integrators
Vision Transformers process images fundamentally differently by dividing them into patches and treating these patches as a sequence of tokens, similar to how Transformers process words in natural language. Through self-attention mechanisms, ViTs learn relationships between any two patches regardless of their spatial separation, enabling them to capture global context and long-range dependencies across entire whole-slide images (WSIs) [14]. This global perspective is particularly valuable in pathology for understanding tissue architecture, tumor-stroma interactions, and spatial relationships between distant histological structures.
Unlike CNNs, ViTs have minimal built-in inductive biases about images and instead learn relevant visual patterns directly from data. This flexibility allows them to develop more sophisticated representations but comes at the cost of requiring substantial training data to generalize effectively. The self-attention mechanism computes weighted sums of all input patches, with weights determined by compatibility between patches, allowing the model to focus on clinically relevant regions while suppressing irrelevant information. For example, ViT-based models have demonstrated superior capability in classifying squamous cell carcinoma (SCC) margins on low-quality histopathological images, achieving 0.928 ± 0.027 accuracy compared to 0.86 ± 0.049 for the highest-performing CNN model (InceptionV3) [16].
Performance Comparison: Quantitative Analysis Across Pathology Tasks
Table 1: Performance comparison of CNN vs. ViT architectures across multiple pathology applications
| Pathology Task | Dataset | Best CNN Model (Performance) | Best ViT Model (Performance) | Performance Delta |
|---|---|---|---|---|
| SCC Margin Classification | Low-quality histopathological images | InceptionV3 (Accuracy: 0.860 ± 0.049; AUC: 0.837 ± 0.029) | Custom ViT (Accuracy: 0.928 ± 0.027; AUC: 0.927 ± 0.028) | +7.8% Accuracy, +9.0% AUC [16] |
| Breast Cancer Lymph Node Micrometastasis | BLCN-MiD & Camelyon (4× magnification) | ResNet34 | rMetaTrans (Optimized ViT) | +3.67-6.96% across metrics [17] |
| Dental Image Analysis | 21-study systematic review | Various CNNs | ViT-based models | ViT superior in 58% of studies [18] |
| Colorectal Cancer Classification | EBHI dataset (200×) | Multiple CNNs | Feature fusion with self-attention | 99.68% accuracy [19] |
| Melanoma Diagnosis | ISIC datasets | Ensemble CNNs | CNN-ViT ensemble | 95.25% accuracy [20] |
Table 2: Data efficiency and computational requirements comparison
| Characteristic | CNNs | Vision Transformers |
|---|---|---|
| Data Efficiency | Perform well with limited annotated data [14] | Require large-scale data for effective training [14] |
| Computational Demand | Lower computational requirements [14] | Higher computational complexity during training [17] |
| Training Speed | Faster training cycles | Longer training times [18] |
| Inference Speed | Optimized for deployment, suitable for edge devices [14] | Can be optimized through architectural modifications [17] |
| Pretraining Requirements | ImageNet transfer learning effective | Benefit from domain-specific pretraining [8] |
Foundation Models vs. Traditional Transfer Learning: Methodological Divide
The architectural evolution from CNNs to ViTs coincides with a methodological shift from traditional transfer learning to foundation models. Traditional transfer learning typically involves pretraining a model on a large-scale natural image dataset (e.g., ImageNet), then fine-tuning the weights on a smaller target pathology dataset. This approach leverages generalized visual features but suffers from domain shift when natural images differ substantially from histopathological images [19]. For example, a study on colorectal cancer classification applied domain-specific transfer learning using CNNs pretrained on intermediate histopathological datasets rather than natural images, enhancing feature relevance for the target domain [19].
In contrast, pathology foundation models are pretrained directly on massive histopathology datasets using self-supervised learning, capturing domain-specific morphological patterns. A prominent example is TITAN (Transformer-based pathology Image and Text Alignment Network), a multimodal whole-slide foundation model pretrained on 335,645 whole-slide images via visual self-supervised learning and vision-language alignment [8]. This approach learns general-purpose slide representations that transfer effectively across diverse clinical tasks without task-specific fine-tuning. Foundation models address the data scarcity challenge in pathology by learning from vast unlabeled datasets, then applying this knowledge to downstream tasks with minimal labeled examples.
Experimental evidence demonstrates that foundation models significantly outperform traditional transfer learning approaches, particularly in low-data regimes and rare disease contexts. Virchow2, a pathology foundation model, delivered the highest performance across multiple tasks from TCGA, CPTAC, and external benchmarks compared to both general vision models and traditional transfer learning approaches [21]. Similarly, TITAN outperformed supervised baselines and existing slide foundation models in cancer subtyping, biomarker prediction, outcome prognosis, and slide retrieval tasks [8].
Diagram 1: Workflow comparison between traditional transfer learning and foundation model approaches in computational pathology. The foundation model paradigm leverages self-supervised pretraining on large-scale pathology-specific datasets, enabling zero-shot application or minimal fine-tuning, while traditional approaches require extensive domain adaptation from natural images.
Experimental Protocols: Methodological Insights from Key Studies
ViT for Low-Quality Histopathological Images
Objective: To evaluate Vision Transformers for squamous cell carcinoma (SCC) margin classification on low-quality histopathological images from resource-limited settings [16].
Dataset: Comprised histopathological slides from 50 patients with SCC (17 well-differentiated, 15 moderately differentiated, 18 invasive SCC) from Jimma University Medical Center in Ethiopia, including 345 normal tissue images and 483 tumor images designated as margin positive.
Preprocessing: Original high-resolution images (2048 × 1536 pixels) were resized to 224 × 224 pixels. Data augmentation techniques included flipping, scaling, and rotation to increase dataset diversity and prevent overfitting.
Model Architecture: Custom ViT architecture employing transfer learning approach with additional flattening, batch normalization, and dense layers. Implemented five-fold cross-validation for robust performance estimation.
Evaluation Metrics: Primary metrics included accuracy, area under the curve (AUC), with ablation studies exploring architectural configuration effects.
MetaTrans for Breast Cancer Lymph Node Micrometastasis
Objective: To develop MetaTrans, a novel network combining meta-learning with Transformer blocks for detecting lymph node micro-metastases in breast cancer under limited data conditions [17].
Dataset: Constructed 34-category WSI dataset (MT-MCD) for meta-training, including multi-center small metastasis datasets with both paraffin and frozen sections.
Architecture: Integrated meta-learning with Transformer blocks to address limitations of pure Transformers in capturing fine-grained local details of micro-lesions. Employed tissue-recognition model for regions of interest at low magnification (Model4x) and cell-recognition model for high magnification (Model10x).
Training Strategy: Inspired by pathologists' diagnostic practices, the process captures a field of view at 4× magnification, divides it into 256 × 256 patches, processes them with MetaTrans to generate probability distribution and attention maps within 5 seconds.
Evaluation: Comprehensive cross-dataset and cross-disease validation on BLCN-MiD and Camelyon datasets, comparing against CNN baselines (ResNet18, ResNet34, ResNet50) and vanilla ViT architectures (ViTSmall, ViTBase).
TITAN Foundation Model Pretraining
Objective: To develop TITAN, a multimodal whole-slide foundation model for general-purpose slide representation learning in histopathology [8].
Pretraining Data: Mass-340K dataset comprising 335,645 WSIs across 20 organ types with diverse stains, tissue types, and scanner types, plus 182,862 medical reports.
Three-Stage Pretraining:
- Vision-only unimodal pretraining: Using iBOT framework on region crops (8,192 × 8,192 pixels at 20× magnification)
- ROI-level cross-modal alignment: With 423k pairs of ROIs and synthetic captions generated using PathChat
- WSI-level cross-modal alignment: With 183k pairs of WSIs and clinical reports
Architecture Innovations: Extended attention with linear bias (ALiBi) to 2D for long-context extrapolation; constructed input embedding space by dividing each WSI into non-overlapping patches of 512 × 512 pixels at 20× magnification; used CONCHv1.5 for patch feature extraction.
Evaluation Tasks: Diverse clinical applications including cancer subtyping, biomarker prediction, outcome prognosis, slide retrieval, rare cancer retrieval, cross-modal retrieval, and pathology report generation in zero-shot settings.
Diagram 2: Complementary strengths of CNNs and Vision Transformers in computational pathology, showing how emerging hybrid architectures integrate benefits from both approaches for enhanced clinical applications.
Table 3: Key research reagents and computational resources for pathology AI research
| Resource Category | Specific Examples | Function/Application |
|---|---|---|
| Public Datasets | Camelyon (lymph node metastases), TCGA (multi-cancer), CPTAC (proteogenomic), NCT-CRC-HE-100K (colorectal cancer) | Benchmarking, pretraining, and validation of models across different cancer types and tasks [17] [21] |
| Pretrained Models | CONCH (histopathology patch encoder), Virchow2 (pathology foundation model), TITAN (whole-slide foundation model) | Transfer learning, feature extraction, and baseline comparisons for development [8] [21] |
| Evaluation Frameworks | Five-fold cross-validation, external validation datasets, ablation studies | Robust performance assessment and generalization testing [16] [17] |
| Computational Infrastructure | High-memory GPUs (for processing whole-slide images), distributed training systems | Handling computational demands of transformer architectures and large whole-slide images [8] |
| Data Augmentation Tools | Flipping, rotation, scaling, stain normalization, synthetic data generation | Addressing data scarcity and improving model generalization [16] [19] |
The evolution from CNNs to Vision Transformers in computational pathology represents more than a mere architectural shift—it embodies a fundamental transformation in how artificial intelligence models perceive and interpret histopathological images. CNNs remain highly valuable for resource-constrained environments, edge deployments, and tasks where local feature extraction is paramount, while ViTs excel in whole-slide analysis, global context modeling, and foundation model applications. The emerging consensus favors hybrid approaches that integrate the complementary strengths of both architectures, such as ConvNeXt, Swin Transformers, and MetaTrans [14] [17].
The parallel transition from traditional transfer learning to pathology-specific foundation models addresses critical limitations in domain adaptation and data efficiency. Foundation models like TITAN and Virchow2 demonstrate that self-supervised pretraining on massive histopathology datasets produces more versatile and generalizable representations than ImageNet-based transfer learning [8] [21]. For researchers and drug development professionals, strategic model selection should consider not only architectural differences but also the pretraining paradigm, with foundation models increasingly becoming the preferred approach for their superior performance in few-shot and zero-shot settings.
As computational pathology continues to evolve, the integration of multimodal data (imaging, genomics, clinical reports) through transformer-based architectures represents the most promising direction for developing comprehensive AI diagnostic systems that can meaningfully assist pathologists and accelerate drug development workflows.
The field of computational pathology is undergoing a profound transformation, driven by a quantum leap in the scale of training data. The transition from models trained on thousands of Whole-Slide Images (WSIs) to those trained on hundreds of thousands or even millions represents a pivotal shift from task-specific algorithms toward general-purpose foundation models. This evolution mirrors the trajectory seen in natural language processing, where large-scale pretraining has unlocked unprecedented capabilities. In pathology, foundation models are deep neural networks pretrained on massive collections of histology image fragments without specific human labels, learning to understand cellular patterns, tissue architecture, and staining variations across diverse organs and diseases [22]. Unlike traditional transfer learning—which often adapts models pretrained on natural images to specific medical tasks with limited data—foundation models are pretrained directly on vast histopathology datasets, capturing the intrinsic morphological diversity of human tissue at scale [22]. This paradigm shift enables models to serve as versatile visual backbones that can be efficiently adapted to numerous clinical and research applications with minimal fine-tuning.
Quantitative Comparison: Traditional vs. Foundation Model Approaches
Table 1: Comparison of Data Scale and Model Architectures
| Model / Approach | Training Data Scale | Model Architecture | Pretraining Strategy |
|---|---|---|---|
| Traditional Transfer Learning | Thousands to tens of thousands of WSIs [23] [24] | Convolutional Neural Networks (CNNs) [24] | Supervised learning on specific tasks [24] |
| TITAN | 335,645 WSIs across 20 organ types [8] | Vision Transformer (ViT) [8] | Self-supervised learning (iBOT) & multimodal alignment [8] |
| Prov-GigaPath | 171,189 WSIs (1.3 billion image patches) [22] | GigaPath (using LongNet dilated attention) [22] | Self-supervised learning on gigapixel images [22] |
| UNI | 100,000+ WSIs (100 million patches) [22] | Vision Transformer [22] | Self-supervised learning for universal representation [22] |
Performance Comparison Across Clinical Tasks
Table 2: Performance Metrics on Diagnostic and Prognostic Tasks
| Model / Approach | Cancer Subtyping Accuracy | Rare Disease Retrieval | Zero-shot Classification | Biomarker Prediction |
|---|---|---|---|---|
| Traditional CNN (Weakly Supervised) | 0.908 micro-accuracy (image-level) [24] | Not reported | Not applicable | Not reported |
| TITAN | Outperforms ROI and slide foundation models across multiple subtyping tasks [8] | Superior performance in rare cancer retrieval [8] | Enables zero-shot classification via vision-language alignment [8] | Outperforms supervised baselines [8] |
| Prov-GigaPath | State-of-the-art in 25/26 clinical tasks, including 9 cancer types [22] | Not specifically reported | Enables zero-shot classification from clinical descriptions [22] | Predicts genetic alterations (e.g., MSI status) from H&E [22] |
| Virchow | AUC 0.95 for tumor detection across 9 common and 7 rare cancers [22] | Demonstrates impressive generalization for rare cancers [22] | Not specifically reported | Outperforms organ-specific clinical models with fewer labeled data [22] |
Experimental Protocols and Methodologies
Foundation Model Pretraining Workflow
Foundation Model Pretraining Pathway
The TITAN model exemplifies the sophisticated methodologies employed in modern pathology foundation models. The pipeline begins with Whole-Slide Images (WSIs) being processed through a patch embedding layer, typically using established patch encoders like CONCH, to extract meaningful feature representations [8]. These features are spatially arranged in a two-dimensional grid that preserves the architectural context of the tissue [8]. The pretraining proceeds through three progressive stages: (1) vision-only self-supervised learning using the iBOT framework that employs knowledge distillation and masked image modeling; (2) cross-modal alignment with fine-grained morphological descriptions generated from 423,000 synthetic captions; and (3) slide-level alignment with 183,000 pathology reports [8]. This multistage approach enables the model to learn both visual semantics and their correspondence with clinical language, unlocking capabilities in zero-shot reasoning and cross-modal retrieval.
Traditional Weakly-Supervised Approach
Traditional Weakly-Supervised Learning Pathway
Traditional approaches rely on weakly-supervised frameworks that automatically extract labels from pathology reports to train convolutional neural networks. The Semantic Knowledge Extractor Tool (SKET) employs an unsupervised hybrid approach combining rule-based systems with pretrained machine learning models to derive semantically meaningful concepts from free-text diagnostic reports [24]. These automatically generated labels then train a Multiple Instance Learning CNN framework that processes individual patches from WSIs and aggregates predictions using attention pooling to produce slide-level classifications [24]. While this approach eliminates the need for manual annotations and leverages existing clinical data, it remains constrained by its focus on specific tasks and limited ability to generalize beyond its training distribution.
Table 3: Key Research Reagents and Computational Resources
| Resource Category | Specific Examples | Function in Research |
|---|---|---|
| Whole-Slide Scanners | Leica Aperio AT2/GT450, Hamamatsu NanoZoomer S360, Philips UFS, 3DHistech Pannoramic 1000 [25] | Digitize glass slides into high-resolution whole-slide images for computational analysis |
| Patch Encoders | CONCH, CONCHv1.5 [8] | Encode histopathology regions-of-interest into feature representations for slide-level processing |
| Computational Frameworks | Vision Transformers (ViT), GigaPath with LongNet dilated attention [22] | Process long sequences of patch embeddings from gigapixel WSIs while capturing global context |
| Self-Supervised Learning Methods | iBOT, masked autoencoders (MAE), DINO contrastive learning [8] [22] | Pretrain models without human annotations by solving pretext tasks like masked image modeling |
| Multimodal Datasets | Pathology reports, synthetic captions, transcriptomics data [8] [22] | Provide complementary supervisory signals for vision-language alignment and multimodal reasoning |
| Quality Control Tools | Focus assessment, artifact detection, missing tissue identification [25] [26] | Ensure digital slide quality and identify scanning errors that could compromise model performance |
Discussion: Implications for Research and Drug Development
The leap from thousands to millions of whole-slide images represents more than a quantitative scaling—it marks a qualitative transformation in how computational pathology approaches pattern recognition and diagnostic reasoning. Foundation models pretrained at this scale demonstrate emergent capabilities, including zero-shot classification, rare disease retrieval, and molecular pattern inference directly from H&E stains [8] [22]. For research and drug development, these advances offer compelling opportunities: identifying subtle morphological biomarkers invisible to the human eye, predicting treatment response through integrated analysis of histology with clinical and genomic data, and accelerating drug discovery by revealing novel genotype-phenotype correlations [22].
The integration of multimodal data streams represents perhaps the most promising frontier. Models like THREADS now align histological images with RNA-seq expression profiles and DNA data, creating bridges between tissue morphology and molecular signatures [22]. Similarly, approaches like MIFAPS integrate MRI, whole-slide images, and clinical data to predict pathological complete response in breast cancer [22]. For pharmaceutical researchers, these capabilities enable more precise patient stratification, biomarker discovery, and understanding of drug mechanisms across tissue contexts.
However, this paradigm shift also introduces new challenges. The computational resources required for training foundation models are substantial, often requiring tens of thousands of GPU hours [22]. Data standardization remains critical, as variations in staining protocols, scanner models, and tissue preparation can significantly impact model performance [23] [25]. Importantly, the transition to foundation models does not eliminate the need for domain expertise—rather, it repositions pathologists as interpreters of model outputs and validators of clinical relevance [22].
The scaling of training data from thousands to millions of whole-slide images has catalyzed a fundamental shift from task-specific models to general-purpose foundation models in computational pathology. This transition has demonstrated unequivocal benefits in diagnostic accuracy, generalization to rare conditions, and multimodal reasoning capabilities. While traditional transfer learning approaches remain viable for focused applications with limited data, foundation models offer a more versatile and powerful paradigm for organizations with access to large-scale data and computational resources. As these models continue to evolve, they promise to deepen our understanding of disease biology and accelerate the development of targeted therapies through their ability to discern subtle morphological patterns and their correlations with molecular features and clinical outcomes.
The development of computational pathology tools has been historically constrained by the limited availability of large-scale annotated histopathology datasets. Traditional transfer learning from natural image domains (e.g., ImageNet) presents significant limitations due to domain shift issues, as histopathology images exhibit fundamentally different characteristics including complex tissue structures, specific staining patterns, and substantially higher resolution. Self-supervised learning (SSL) has emerged as a transformative paradigm that leverages inherent patterns within unannotated data to learn robust, transferable representations, effectively addressing annotation bottlenecks in medical imaging [27] [28].
Within SSL, two predominant frameworks have demonstrated remarkable success in histopathology applications: contrastive learning and masked image modeling. These approaches differ fundamentally in their learning objectives, architectural requirements, and performance characteristics across various computational pathology tasks. This comparative analysis examines their methodological principles, experimental performance, and implementation considerations within the broader context of foundation model development for histopathology, providing researchers with evidence-based guidance for selecting appropriate paradigms for specific clinical and research applications.
Methodological Foundations
Contrastive Learning Frameworks
Contrastive learning operates on the principle of discriminative representation learning by maximizing agreement between differently augmented views of the same image while pushing apart representations from different images. In histopathology, this approach has been extensively adapted to handle the unique characteristics of whole-slide images (WSIs), including their gigapixel sizes and hierarchical tissue structures [29] [30].
The core objective function typically follows the Noise Contrastive Estimation (InfoNCE) framework, which aims to identify positive pairs (different views of the same histopathology patch) among negative samples (views from different patches). Key implementations in histopathology include:
- SimCLR (Simplified Contrastive Learning of Representations): Employed on collections of 57 histopathology datasets without labels, demonstrating that combining multi-organ datasets with varied staining and resolution properties improves learned feature quality [29].
- MoCo v2 (Momentum Contrast v2): Used for in-domain pretraining on colon adenocarcinoma cohorts from TCGA, significantly outperforming ImageNet pretraining on metastasis detection tasks [28].
- DINO (self-DIstillation with NO labels): Leverages vision transformers (ViTs) through self-distillation without labels, using similarity matching between global and local crop embeddings via cross-entropy loss [28].
Masked Image Modeling Frameworks
Masked image modeling (MIM) draws inspiration from masked language modeling in natural language processing (e.g., BERT), where the model learns to predict masked portions of the input data based on contextual information. For histopathology images, this approach forces the model to develop a comprehensive understanding of tissue microstructure and spatial relationships [31] [28].
The iBOT framework (image BERT pre-training with Online Tokenizer) has emerged as a particularly effective MIM implementation for histopathology, combining masked patch modeling with online tokenization. Key characteristics include:
- Dual self-distillation objectives: Simultaneously learns both low-level histomorphological details (through masked patch reconstruction) and high-level visual semantics (through class token distillation) [31] [28].
- Online tokenizer: Dynamically generates training targets through a momentum teacher network, avoiding the need for predefined visual vocabularies.
- Pan-cancer representation learning: Effectively captures morphological patterns across diverse cancer types when trained on large-scale datasets (e.g., 40+ million images from 16 cancer types) [31].
Table 1: Core Methodological Differences Between SSL Paradigms
| Aspect | Contrastive Learning | Masked Image Modeling |
|---|---|---|
| Learning Objective | Discriminate between similar and dissimilar image pairs | Reconstruct masked portions of input images |
| Primary Signal | Instance discrimination | Contextual prediction |
| Data Augmentation | Heavy reliance on carefully designed augmentations | Less dependent on complex augmentations |
| Architecture | Compatible with CNNs and ViTs | Primarily optimized for Vision Transformers |
| Representation Level | Emphasis on global semantics | Balances local texture and global structure |
Experimental Performance Comparison
Benchmarking Studies and Direct Comparisons
Comprehensive evaluations across diverse histopathology tasks consistently demonstrate the advantages of in-domain SSL pretraining over traditional ImageNet transfer learning. However, significant performance differences exist between contrastive and MIM approaches depending on task characteristics and data regimes.
The iBOT framework, as a leading MIM implementation, has demonstrated state-of-the-art performance across 17 downstream tasks spanning seven cancer indications, including weakly-supervised WSI classification and patch-level tasks. Specifically, iBOT pretrained on pan-cancer datasets outperformed both ImageNet pretraining and MoCo v2 (a contrastive approach) on tasks including microsatellite instability (MSI) prediction, homologous recombination deficiency (HRD) classification, cancer subtyping, and overall survival prediction [28].
Notably, MIM approaches exhibit particularly strong performance in low-data regimes, maintaining robust representation quality even with limited fine-tuning examples. This property is especially valuable in histopathology, where annotated datasets for rare cancers or molecular subtypes are often small [28]. Contrastive methods, while generally effective, show greater performance degradation when pretraining datasets exhibit significant class imbalance - a common scenario in real-world histopathology collections [27].
Scaling Properties and Data Efficiency
The development of foundation models in histopathology depends critically on understanding how performance scales with model size, data volume, and data diversity. Evidence suggests that MIM approaches exhibit favorable scaling properties compared to contrastive methods:
- Model Scaling: Vision Transformers pretrained with iBOT demonstrate consistent performance improvements as model size increases from 22 million to 307 million parameters, particularly when coupled with larger pretraining datasets [28].
- Data Scaling: MIM performance improves monotonically with increased pretraining data size and diversity. iBOT pretrained on 43 million histology images from 16 cancer types outperformed versions trained on smaller, organ-specific cohorts [31].
- Multimodal Extension: MIM representations serve as effective foundations for vision-language models like TITAN (Transformer-based pathology Image and Text Alignment Network), enabling zero-shot classification and cross-modal retrieval without task-specific fine-tuning [8].
Table 2: Performance Comparison Across Histopathology Tasks
| Task Category | Contrastive Learning (SimCLR/MoCo) | Masked Image Modeling (iBOT) | Evaluation Metric |
|---|---|---|---|
| Slide-Level Classification | 28% improvement over ImageNet [29] | Outperforms MoCo v2 by 3-8% [28] | F1 Score / AUROC |
| Patch-Level Classification | Comparable to ImageNet pretraining [29] | Significant improvements on nuclear segmentation and classification | Accuracy |
| Molecular Prediction | Moderate performance on MSI/HRD prediction | State-of-the-art on pan-cancer mutation prediction [28] | AUROC |
| Survival Prediction | Limited demonstrations | Strong performance in multi-cancer evaluation [28] | C-Index |
| Few-Shot Learning | Moderate transferability | Excellent performance with limited labels [28] | Accuracy |
Implementation Considerations
Technical Requirements and Workflows
Implementing SSL frameworks in histopathology requires specialized computational infrastructure and data processing pipelines. The following diagram illustrates a generalized workflow for MIM-based foundation model development:
Data Preprocessing Requirements:
- Patch Extraction: WSIs are divided into smaller patches (typically 256×256 or 512×512 pixels at 20× magnification) [8] [28].
- Color Normalization: Addresses staining variations across institutions using methods like Macenko or Vahadane normalization.
- Feature Grid Construction: For slide-level modeling, patches are arranged in 2D spatial grids preserving tissue topology [8].
Computational Infrastructure:
- Hardware: Multiple high-end GPUs (e.g., A100 or H100) with substantial VRAM (≥80GB).
- Training Time: Ranges from days to weeks depending on dataset size (millions to hundreds of millions of patches).
- Storage: Large-scale distributed storage systems capable of handling petabyte-scale whole-slide image repositories.
Essential Research Reagents
Table 3: Key Research Reagents for SSL in Histopathology
| Resource Category | Specific Examples | Function and Application |
|---|---|---|
| Pretrained Models | iBOT (ViT-Base/Large), UNI, CTransPath, CONCH | Foundation models for transfer learning and feature extraction [31] [8] [32] |
| Histopathology Datasets | TCGA (The Cancer Genome Atlas), Camelyon, NCT-CRC-HE-100K, Mass-100K | Large-scale WSI collections for pretraining and benchmarking [29] [32] |
| Software Libraries | TIAToolbox, VISSL, PyTorch, Whole-Slide Data Loaders | Data preprocessing, model implementation, and evaluation pipelines |
| Evaluation Frameworks | HistoPathExplorer, PMCB (Pathology Model Benchmark) | Performance tracking across multiple tasks and datasets [33] |
Integration with Foundation Model Development
The evolution from specialized models to general-purpose foundation models represents a paradigm shift in computational pathology. Both contrastive learning and MIM contribute uniquely to this transition:
Contrastive Learning's Role:
- Established the viability of in-domain pretraining for histopathology
- Demonstrated that multi-institutional datasets with diverse staining protocols improve robustness [29]
- Provided simple yet effective frameworks for representation learning with limited annotations
MIM's Advantages for Foundation Models:
- Superior scalability with model and data size [28]
- Natural extension to multimodal learning (e.g., vision-language models) [8]
- Stronger transfer performance across diverse tissue types and disease categories
- Enhanced few-shot and zero-shot capabilities critical for rare cancer applications
The UNI model exemplifies this transition, having been pretrained on >100 million images from >100,000 H&E-stained WSIs across 20 tissue types using DINOv2 (a MIM-inspired framework). UNI demonstrates remarkable versatility across 34 clinical tasks, including resolution-agnostic tissue classification and few-shot cancer subtyping for up to 108 cancer types in the OncoTree system [32].
The following diagram illustrates how SSL paradigms integrate into the broader foundation model ecosystem:
The comparative analysis of self-supervised learning paradigms in histopathology reveals a clear trajectory toward masked image modeling as the foundational approach for next-generation computational pathology tools. While contrastive learning established the critical principle that in-domain pretraining surpasses transfer learning from natural images, MIM methods like iBOT demonstrate superior performance across diverse tasks, better scaling properties, and stronger generalization to rare cancers and low-data scenarios.
Several emerging trends will shape future developments in this field:
- Multimodal Integration: Combining visual self-supervision with pathology reports and molecular profiles, as exemplified by TITAN, enables more comprehensive representation learning and zero-shot capabilities [8].
- Scaled Architectures: Vision Transformers with hundreds of millions of parameters, pretrained on increasingly diverse histopathology datasets (approaching the petabyte scale), continue to push performance boundaries [32].
- Federated Learning: Self-supervised approaches are being adapted for distributed training across institutions while preserving data privacy, though challenges remain in identifying quality issues without direct data inspection [34].
For researchers and drug development professionals selecting SSL approaches for histopathology applications, MIM frameworks currently offer the most promising path for developing robust, generalizable models, particularly when targeting multiple downstream tasks or working with limited annotations. Contrastive methods remain viable for more focused applications with sufficient annotated data for fine-tuning. As foundation models continue to evolve in computational pathology, the integration of SSL with multimodal data and clinical domain knowledge will ultimately bridge the gap between experimental AI capabilities and routine pathological practice.
Implementation in Precision Oncology: Methodologies and Real-World Applications
In computational pathology, the emergence of foundation models represents a paradigm shift from traditional transfer learning approaches. Traditional transfer learning typically involves taking a model pre-trained on a general dataset (like ImageNet) and fine-tuning it on a specific, often limited, pathology dataset. While beneficial, this method remains constrained by its dependency on large, annotated datasets for each new task and its limited ability to integrate diverse data types. Foundation models, pre-trained on vast and diverse datasets using self-supervised learning, offer a more powerful alternative. They provide generalized representations that can be adapted to numerous downstream tasks with minimal task-specific data, thereby addressing key limitations of traditional methods [8] [35].
Within this context, a critical architectural division has emerged: uni-modal (vision-only) models and multi-modal (vision-language) models. Uni-modal models process exclusively image data, focusing on learning rich visual representations from histopathology slides. In contrast, multi-modal models learn from both images and associated textual data (such as pathology reports), creating a shared representation space that enables a broader range of capabilities, including cross-modal retrieval and zero-shot reasoning [8]. This guide provides a comparative analysis of these two model archetypes, focusing on their application within computational pathology research and drug development.
Comparative Performance Analysis
Evaluations across diverse clinical tasks reveal distinct performance profiles for uni-modal and multi-modal foundation models. The table below summarizes quantitative findings from key studies, highlighting the strengths of each archetype.
Table 1: Performance Comparison of Uni-Modal vs. Multi-Modal Foundation Models in Pathology Tasks
| Model Archetype | Example Model | Key Performance Metrics | Optimal Use Cases |
|---|---|---|---|
| Uni-Modal (Vision-Only) | TITAN-V (Vision-only variant) | High performance in slide-level tasks like cancer subtyping and biomarker prediction [8]. | Standard visual classification, prognosis prediction, tasks where only image data is available. |
| Multi-Modal (Vision-Language) | TITAN (Full vision-language model) | Outperforms slide foundation models in few-shot and zero-shot classification; enables cross-modal retrieval and pathology report generation [8]. | Low-data regimes, rare disease retrieval, tasks requiring integration of visual and textual information. |
The TITAN model exemplifies the power of multi-modal learning. In rigorous benchmarking, it demonstrated superior performance over both region-of-interest (ROI) and slide-level foundation models across multiple machine learning settings, including linear probing, few-shot learning, and zero-shot classification [8]. This is particularly valuable for rare diseases and low-data scenarios, where traditional models struggle. For instance, multi-modal models can retrieve similar cases based on either an image query or a text description of morphological findings, a capability beyond the reach of vision-only systems [8].
However, vision-only models remain highly effective for well-defined visual tasks with sufficient training data. They avoid the complexity and computational overhead of processing multiple modalities and can achieve state-of-the-art results in tasks such as cancer subtyping and outcome prognosis [8]. The choice of archetype, therefore, depends heavily on the specific clinical or research application, data availability, and the need for linguistic understanding.
Experimental Protocols and Methodologies
The development and validation of foundation models in pathology require rigorous and standardized experimental protocols. The following workflow outlines a typical methodology for pre-training and evaluating a multi-modal model like TITAN.
Diagram 1: Foundation Model Pre-training Workflow
Detailed Experimental Methodology
The experimental protocol for a model like TITAN involves a multi-stage pre-training pipeline, as illustrated above [8]:
Data Curation and Preprocessing:
- Vision-Only Pre-training: A large dataset of Whole-Slide Images (WSIs) is required. For TITAN, this involved 335,645 WSIs (Mass-340K dataset) across 20 organ types to ensure diversity [8].
- Multi-Modal Pre-training: This stage requires paired image-text data. This includes both synthetic fine-grained captions for Regions of Interest (ROIs)—423,122 captions generated via a generative AI copilot for pathology—and 182,862 slide-level pathology reports [8].
- ROI Feature Extraction: WSIs are divided into non-overlapping patches (e.g., 512x512 pixels at 20x magnification). A pre-trained patch encoder (e.g., CONCH) is used to extract a feature vector for each patch, creating a 2D feature grid that preserves spatial relationships [8].
Model Pre-training:
- Stage 1 (Uni-Modal, Vision-Only): The model (e.g., a Vision Transformer) is trained on the 2D feature grid using self-supervised learning (SSL) methods like iBOT, which combines masked image modeling and knowledge distillation. This stage teaches the model fundamental histomorphological patterns [8].
- Stage 2 (Multi-Modal, ROI-Text Alignment): The vision model is aligned with text at a fine-grained level using the synthetic ROI captions. This is typically done with contrastive learning, which pulls the representations of matching image-text pairs closer while pushing non-matching pairs apart [8].
- Stage 3 (Multi-Modal, Slide-Report Alignment): The model is further aligned at a broader slide level using the paired WSIs and pathology reports. This step bridges the gap between detailed visual features and overarching diagnostic language [8].
Evaluation and Downstream Tasks:
- The model's general-purpose slide representations are evaluated on diverse clinical tasks without end-to-end fine-tuning ("zero-shot" or with "linear probing"). Key benchmarks include [8]:
- Diagnostic Classification: Cancer subtyping and biomarker prediction.
- Prognosis: Predicting patient outcomes from histology.
- Retrieval: Rare cancer retrieval using image or text queries.
- Generation: Automatic generation of pathology reports from a WSI.
- The model's general-purpose slide representations are evaluated on diverse clinical tasks without end-to-end fine-tuning ("zero-shot" or with "linear probing"). Key benchmarks include [8]:
The Scientist's Toolkit: Key Research Reagents
Successfully developing or applying foundation models in computational pathology requires a suite of key "research reagents." The following table details these essential components.
Table 2: Essential Research Reagents for Pathology Foundation Models
| Item | Function & Importance |
|---|---|
| Large-Scale WSI Datasets | Foundation models require massive, diverse datasets for pre-training. The Mass-340K dataset (335k WSIs) is an example, encompassing multiple organs, stains, and scanner types to learn robust, generalizable features [8]. |
| Paired Image-Text Data | For multi-modal models, high-quality paired data is critical. This includes both synthetic captions for ROIs and real-world pathology reports, enabling the model to link visual patterns with semantic descriptions [8]. |
| Pre-trained Patch Encoder | Models like CONCH convert image patches into feature embeddings. These pre-extracted features form the foundational "vocabulary" for the slide-level transformer model, making training computationally feasible [8]. |
| SSL Algorithms (e.g., iBOT) | Self-supervised algorithms leverage unlabeled data by creating learning signals from the data itself (e.g., reconstructing masked patches). This is the core mechanism for building general visual representations without manual labels [8]. |
| Computational Infrastructure | Training on gigapixel WSIs demands significant resources, including high-memory GPUs and optimized software frameworks (e.g., PyTorch, Transformers), to handle long input sequences and complex model architectures [36] [8]. |
Discussion and Future Directions
The transition from traditional transfer learning to foundation models marks a significant evolution in computational pathology. Uni-modal vision models offer a powerful, direct path for tasks centered purely on image analysis, building on the established principles of deep learning for visual recognition. However, multi-modal vision-language models like TITAN represent a qualitative leap forward. By integrating visual and textual information, they more closely mimic the holistic reasoning process of a pathologist, who correlates microscopic findings with clinical context and descriptive language [8]. This enables novel capabilities such as zero-shot reasoning, cross-modal search, and language-guided interpretation, which are invaluable for drug development in identifying novel biomarkers and stratifying patient populations [37] [38].
Despite their promise, both archetypes face challenges for clinical integration. "Black-box" nature and interpretability issues can hinder clinician trust [36] [37]. Furthermore, multi-modal models introduce additional complexity regarding data privacy, algorithmic bias potentially amplified by biased text reports, and the high computational cost of training and deployment [36] [8]. Future research will focus on improving model interpretability, enhancing generalizability across diverse populations and laboratory protocols, and developing more efficient architectures to make these powerful tools more accessible and trustworthy for routine clinical and research use [36] [39].
This guide provides an objective comparison of three leading foundation models in computational pathology: CONCH, Virchow2, and UNI. Framed within the broader thesis of foundation models versus traditional transfer learning, we detail their technical profiles, performance data, and the experimental protocols used for their evaluation.
Model Specifications and Training Data
The table below summarizes the core architectural and training data specifications for each model.
Table 1: Technical Profiles of CONCH, Virchow2, and UNI
| Feature | CONCH | Virchow2 | UNI |
|---|---|---|---|
| Model Type | Vision-Language (Multimodal) | Vision-Only | Vision-Only |
| Core Architecture | ViT-B (Image Encoder) & Text Encoder [40] | ViT-H (632M) / ViT-G (1.85B) [41] | ViT-L (ViT-Large) [32] |
| Primary Training Algorithm | Contrastive Learning & Captioning (based on CoCa) [40] | DINOv2 with domain adaptations [41] | DINOv2 [32] |
| Training Data Scale | 1.17 million image-caption pairs [40] | 3.1 million WSIs [41] | 100 million images from 100,000+ WSIs (Mass-100K) [32] |
| Key Data Sources | Diverse histopathology images and biomedical text (e.g., PubMed) [40] [42] | 3.1M WSIs from globally diverse institutions; mixed stains (H&E, IHC) [41] | Mass-100K (H&E stains from MGH, BWH, GTEx) [32] |
Comparative Performance Benchmarks
Independent, large-scale benchmarking reveals how these models perform across clinically relevant tasks. The following table summarizes key results.
Table 2: Comparative Model Performance on Downstream Tasks
| Evaluation Task / Metric | CONCH | Virchow2 | UNI | Notes & Source |
|---|---|---|---|---|
| Overall Average AUROC (31 tasks) | 0.71 [4] | 0.71 [4] | 0.68 [4] | Across morphology, biomarkers, prognosis [4] |
| Morphology (Avg. AUROC) | 0.77 [4] | 0.76 [4] | - | - |
| Biomarkers (Avg. AUROC) | 0.73 [4] | 0.73 [4] | - | - |
| Prognosis (Avg. AUROC) | 0.63 [4] | 0.61 [4] | - | - |
| Rare Cancer Detection (AUC) | - | 0.937 (pan-cancer) [43] | Strong scaling to 108 cancer types [32] | Virchow: 7 rare cancers; UNI: OncoTree evaluation [43] [32] |
| Zero-shot NSCLC Subtyping (Accuracy) | 90.7% [40] | - | - | CONCH outperformed other V-L models [40] |
| Data Efficiency | Superior in full-data settings [4] | Strong in low-data scenarios [4] | - | Virchow2 led more tasks with 75-300 training samples [4] |
Experimental Protocols for Benchmarking
The performance data in this guide is largely derived from a comprehensive, independent benchmark study [4]. The detailed methodology is as follows:
- Models Evaluated: 19 foundation models, including CONCH, Virchow2, and UNI.
- Downstream Tasks: 31 binary classification tasks across 5 morphological classifications, 19 biomarker predictions, and 7 prognostic outcome predictions.
- Datasets: 9,528 slides from 6,818 patients with lung, colorectal, gastric, and breast cancers. All cohorts were external and not used in the pretraining of the evaluated models to prevent data leakage.
- Weakly-Supervised Learning Framework:
- Feature Extraction: Each WSI was divided into non-overlapping tissue patches. Patch-level embeddings were extracted using each foundation model without any fine-tuning.
- Slide-Level Aggregation: A Transformer-based multiple instance learning (MIL) aggregator was trained to make slide-level predictions from the set of patch-level embeddings.
- Evaluation: Model performance was assessed using the Area Under the Receiver Operating Characteristic Curve (AUROC) on external test sets. Statistical significance was tested with DeLong's test.
This protocol ensures a fair and clinically relevant comparison by testing the models' ability to produce high-quality, transferable representations for tasks with limited labels.
Model Training Workflows
The diagram below illustrates the core self-supervised learning paradigms used by these foundation models, which enable learning from vast amounts of unlabeled data.
Research Reagent Solutions
The following table details key computational "reagents" – the software models and datasets essential for research in this field.
Table 3: Essential Research Reagents for Computational Pathology
| Reagent / Resource | Type | Primary Function in Research | Example / Source |
|---|---|---|---|
| CONCH Model | Vision-Language Foundation Model | Enables tasks involving images and/or text: zero-shot classification, cross-modal retrieval, captioning [40] [42]. | Available on GitHub [42] |
| Virchow / Virchow2 Model | Vision-Only Foundation Model | Provides state-of-the-art image embeddings for slide-level tasks like pan-cancer detection and biomarker prediction [43]. | - |
| UNI Model | Vision-Only Foundation Model | General-purpose image encoder for diverse tasks; demonstrates scaling laws and few-shot learning capabilities [32]. | - |
| DINOv2 Algorithm | Self-Supervised Learning Framework | Core training method for vision-only models; uses a student-teacher framework with contrastive objectives to learn robust features [41] [32]. | - |
| TCGA (The Cancer Genome Atlas) | Public Dataset | A common benchmark dataset for training and evaluating computational pathology models [5]. | - |
| Multiple Instance Learning (MIL) Aggregator | Machine Learning Model | Aggregates patch-level embeddings from a whole slide image to make a single slide-level prediction, enabling weakly supervised learning [4]. | Transformer-based, ABMIL |
In computational pathology, the emergence of foundation models is shifting the paradigm from traditional, task-specific transfer learning towards adaptation from a single, general-purpose model. This guide objectively compares the performance of core adaptation strategies—fine-tuning, linear probing, and few-shot learning—enabling researchers to select the most effective approach for their specific data and task constraints.
The adaptation of large-scale pre-trained models to downstream tasks is a cornerstone of modern computational pathology. Instead of training a separate model for each task from scratch, the prevailing paradigm involves starting with a shared foundation model and adapting it to a specific objective [44]. These foundation models, trained on broad data using self-supervision at scale, can be adapted to a wide range of downstream tasks [44]. The choice of adaptation strategy involves critical trade-offs between performance, data efficiency, computational cost, and preservation of the model's generalizable features.
The three primary adaptation strategies are:
- Fine-Tuning: After adding a task-specific layer that takes the model's features and outputs a prediction, the entire model's parameters are updated via gradient descent [44]. This can be computationally expensive but is often very effective with sufficient data.
- Linear Probing: This method holds the foundation model fixed and trains only a simple linear classifier on top of its extracted features [44]. It is simple and efficient but relies entirely on the quality of the pre-trained features.
- Few-Shot Learning: This encompasses techniques designed to learn from very few examples. Prominent methods include In-Context Learning (ICL), where models like GPT-3.5 or GPT-4 are given a few input-output examples within a prompt to perform the task without updating their weights [45], and SetFit, which uses a contrastive learning approach to tune sentence transformers with limited data [45].
Comparative Performance Analysis
The following tables summarize the performance of these strategies across different experimental conditions, primarily within computational pathology and related vision-language domains.
Table 1: Comparison of Adaptation Strategies Across Key Metrics
| Adaptation Strategy | Data Efficiency | Computational Cost | Performance Potential | Preservation of Generalization | Ideal Use Case |
|---|---|---|---|---|---|
| Full Fine-Tuning | Low (requires more data) | Very High | High [46] | Can suffer from catastrophic forgetting [46] | Data-abundant scenarios where top-tier accuracy is critical |
| Linear Probing | Medium | Low | Moderate [45] [47] | High (model is frozen) | Efficient transfer learning, initial model evaluation |
| Prompt Tuning / ICL | High (few-shot) | Very Low (inference only) | Good with large models [45] | High | Quick prototyping, black-box models (e.g., GPT-4) |
| Specialized Methods (CLAP, PALP) | High (few-shot) | Low to Medium | State-of-the-art in few-shot [48] [47] | Designed to maintain it [47] | Few-shot adaptation where robustness and no hyperparameter tuning are required |
Table 2: Experimental Results in Pathology and Cross-Domain Few-Shot Learning
| Context | Task | Best Performing Model/Strategy | Comparative Performance | Reference |
|---|---|---|---|---|
| Forensic Pathology (PMI Estimation) | Cross-species classification (porcine to human) | UNI (Vision Transformer) with fine-tuning | 91.63% accuracy on porcine data; 78.95% on human data after transfer (outperformed ResNet50, DenseNet121) [49] | |
| Polish NLP Benchmark | 7 classification tasks (16-shot) | In-Context Learning (GPT-3.5/4) | Best performance among few-shot methods;但仍比在全量数据上微调的模型差 14 percentage points [45] [50] | |
| Polish NLP Benchmark | 7 classification tasks (16-shot) | SetFit | Second-best after ICL, closely followed by Linear Probing [45] | |
| Polish NLP Benchmark | 7 classification tasks (16-shot) | Non-linear Head Fine-Tuning | Worst and most unstable performance [45] | |
| Vision-Language Models | EuroSAT & ImageNet generalization | CLIP-CITE (Full Fine-Tuning) | Achieved balanced performance, enhancing specialization while preserving versatility [46] |
Detailed Experimental Protocols
To ensure reproducibility and provide deeper insight, the methodologies of key cited experiments are detailed below.
Cross-Species PMI Estimation with UNI
Objective: To estimate the postmortem interval (PMI) by effectively transferring knowledge from animal (porcine) models to human samples using a pathology foundation model [49]. Models Evaluated: ResNet50, DenseNet121, SongCi, and the UNI foundation model [49]. Protocol:
- Initial Fine-Tuning: The UNI model, a Vision Transformer (ViT) pre-trained on millions of histology whole-slide images (WSIs), was first fine-tuned on WSIs of porcine liver tissues with known PMIs.
- Cross-Species Transfer: The porcine-fine-tuned model was then further fine-tuned (i.e., transfer learning) on a small dataset of 23 human liver WSIs.
- Interpretability Analysis: Model predictions were visualized at the whole-slide level using probability maps, class maps, and classification proportion histograms to enhance transparency [49]. Key Outcome: The two-stage fine-tuning of the UNI model resulted in a greater than 50% improvement in accuracy on human samples compared to the non-fine-tuned model, demonstrating the efficacy of combining foundation models with cross-species transfer learning [49].
Benchmarking Few-Shot Learning for Classification
Objective: To empirically compare few-shot learning techniques across a range of classification tasks [45]. Methods Evaluated: Fine-tuning, linear probing, SetFit, and in-context learning (ICL) [45]. Models: Various pre-trained commercial (e.g., GPT-3.5, GPT-4) and open-source models [45]. Protocol:
- Benchmark Creation: A benchmark of 7 different classification tasks native to the Polish language was established.
- Few-Shot Training: Each method was evaluated in a 0-shot (no examples) and 16-shot (16 examples per class) setting.
- Performance Comparison: The performance of each few-shot method was compared against a strong baseline: a HerBERT-large model fine-tuned on the entire training dataset [45] [50]. Key Outcome: ICL with large commercial models achieved the best few-shot performance, but a significant gap (14 percentage points) remained compared to full-data fine-tuning, highlighting a limitation of few-shot learning [45].
Enhancing Linear Probing for Few-Shot Learning
Objective: To improve the scalability and performance of linear probing in data-limited scenarios, moving beyond the limitations of few-shot in-context learners [48]. Methods: Standard Linear Probing (LP) vs. Prompt-Augmented Linear Probing (PALP) [48]. Protocol:
- Baseline (Standard LP): A linear classifier is trained on top of frozen features extracted from the foundation model.
- PALP Intervention: PALP introduces a constrained optimization objective during linear probing. This penalty prevents the learned class prototypes from deviating too far from the original, general-purpose prototypes derived from the model's zero-shot capabilities.
- Evaluation: The hybrid PALP method was evaluated on various datasets for its ability to scale with the number of available training samples, a scenario where ICL fails due to input length constraints [48]. Key Outcome: PALP was shown to significantly enhance input representations, closing the performance gap between ICL in data-hungry scenarios and fine-tuning in data-abundant scenarios with minimal training overhead [48].
The Scientist's Toolkit: Research Reagents & Models
Table 3: Essential Foundation Models and Computational Tools for Pathology Research
| Name | Type / Category | Primary Function in Research | Key Feature / Architecture |
|---|---|---|---|
| UNI / UNI v2 [51] [49] | Pathology Foundation Model (Vision) | General-purpose feature extraction from H&E and IHC whole-slide images for tasks like classification and survival prediction. | ViT-H/14-reg8; Trained on 200M+ images from 350k+ WSIs [51]. |
| CONCH [5] | Pathology Foundation Model (Vision-Language) | Joint understanding of pathology images and text; enables tasks like text-guided retrieval and report generation. | ViT-B; Trained on 1.17M+ image-caption pairs from PubMed [5]. |
| Virchow v2 [5] | Pathology Foundation Model (Vision) | Patch-level embedding for downstream prediction tasks in oncology. | ViT-H; Trained on 3.1M WSIs from MSKCC [5]. |
| Prov-GigaPath [5] | Pathology Foundation Model (Vision) | Scalable feature extraction for large-scale computational pathology studies. | ViT-G; Trained on 1.3B patches from 171k WSIs [5]. |
| CLIP [46] | General Vision-Language Model | Benchmark model for adapting VLMs to specialized domains with limited data. | Contrastive pre-training on image-text pairs from the web [46]. |
| CLAP Method [47] | Adaptation Algorithm | Enhances linear probing for few-shot learning in VLMs without per-task hyperparameter tuning. | Constrains deviations from zero-shot prototypes [47]. |
| PALP Method [48] | Adaptation Algorithm | A hybrid model that scales beyond the few-shot limit of ICL by combining prompting with linear probing. | Augmented Lagrangian Multiplier optimization [48]. |
Workflow and Strategy Diagrams
Adaptation Strategy Decision Workflow
The following diagram outlines a logical pathway for selecting the most appropriate adaptation strategy based on data availability and task requirements.
Cross-Species Transfer Learning Protocol
This diagram visualizes the two-stage fine-tuning protocol used for PMI estimation, a method that can be generalized to other data-scarce domains.
The prediction of genetic alterations from routine hematoxylin and eosin (H&E)-stained pathology slides represents a transformative application of artificial intelligence in computational pathology. This capability has profound implications for precision medicine, potentially offering a cost-effective and rapid alternative to extensive molecular testing while preserving valuable tissue for comprehensive genomic profiling [52] [53]. The evolution of this field has followed two distinct methodological pathways: traditional transfer learning approaches and emerging foundation models.
Traditional transfer learning typically involves convolutional neural networks (CNNs) pretrained on natural image datasets like ImageNet, which are subsequently fine-tuned on histopathology images for specific biomarker prediction tasks [52]. While this approach has demonstrated promise, it faces fundamental limitations in generalizability and performance, particularly for rare cancer types or biomarkers with subtle morphological correlates [43].
In contrast, pathology foundation models are trained via self-supervised learning on massive, diverse datasets of whole slide images (WSIs), capturing a broad spectrum of histological patterns across tissues, stains, and disease states [43] [54]. These models generate versatile feature representations that transfer robustly to multiple downstream tasks with minimal fine-tuning, potentially overcoming key limitations of traditional approaches [8] [43].
This comparison guide objectively evaluates the performance and methodological considerations of both paradigms, providing researchers and drug development professionals with experimental data and protocols to inform their computational pathology strategies.
Performance Comparison: Foundation Models Versus Traditional Approaches
Quantitative Benchmarking Across Biomarker Types
Table 1: Performance comparison of foundation models versus traditional approaches for biomarker prediction
| Biomarker | Cancer Type | Foundation Model | Traditional Approach | Performance Metric |
|---|---|---|---|---|
| EGFR mutation | Lung adenocarcinoma | EAGLE (AUC: 0.847-0.890) [53] | CNN with weak supervision (AUC: 0.826) [53] | Area Under Curve (AUC) |
| Pan-cancer detection | Multiple cancers | Virchow (AUC: 0.950) [43] | Tissue-specific models | Area Under Curve (AUC) |
| Microsatellite Instability (MSI) | Colorectal cancer | PLUTO-4G [54] | Swin-T transformer [52] | Balanced Accuracy |
| PD-L1 expression | Breast cancer | - | CNN (AUC: 0.85-0.93) [55] | Area Under Curve (AUC) |
| Multiple IHC biomarkers | Gastrointestinal cancers | - | Deep learning models (AUC: 0.90-0.96) [56] | Area Under Curve (AUC) |
Performance in Rare Cancer and Low-Data Scenarios
Table 2: Performance on rare cancers and data-efficient learning
| Evaluation Scenario | Foundation Model | Traditional Approach | Key Findings |
|---|---|---|---|
| Rare cancer detection (7 cancer types) | Virchow (AUC: 0.937) [43] | Tissue-specific models | Foundation models maintain high performance on rare cancers with limited training data |
| Data efficiency | PLUTO-4G [54] | CNN with ImageNet pretraining | Superior performance with limited annotated examples |
| Cross-institutional generalization | TITAN (zero-shot capability) [8] | Requires extensive retraining/adaptation | Maintains performance without fine-tuning on new datasets |
| Metastatic specimens | EAGLE (AUC: 0.75) [53] | - | Reduced but acceptable performance on challenging specimens |
Experimental Protocols and Methodologies
Foundation Model Training Paradigm
The development of pathology foundation models follows a multi-stage self-supervised learning approach optimized for histopathological data:
Large-Scale Pretraining Foundation models like Virchow, PLUTO-4, and TITAN are pretrained on massive WSI datasets comprising 100,000 to over 500,000 slides from diverse institutions, tissue types, and staining protocols [43] [54]. The training employs self-supervised algorithms such as DINOv2, which leverages global and local tissue regions to learn robust morphological representations without manual annotations [43] [54].
Architectural Considerations Modern foundation models utilize Vision Transformer (ViT) architectures with 632 million to over 1 billion parameters, substantially larger than traditional CNNs [43] [54]. The PLUTO-4 family introduces both compact (PLUTO-4S) and frontier-scale (PLUTO-4G) models, incorporating architectural innovations like FlexiViT backbones with Rotary Positional Embeddings for handling multi-scale histopathological features [54].
Multimodal Integration Advanced foundation models like TITAN incorporate multimodal pretraining by aligning visual features with corresponding pathology reports and synthetic captions, enabling cross-modal retrieval and zero-shot classification capabilities [8].
Traditional Transfer Learning Protocol
Traditional approaches for biomarker prediction follow a well-established weakly supervised learning pipeline:
Feature Extraction WSIs are divided into smaller tiles (typically 256×256 or 512×512 pixels at 20× magnification), with non-tissue regions filtered out [52]. Each tile is processed through a CNN pretrained on ImageNet (e.g., ResNet-50) to extract feature representations [52] [56].
Multiple Instance Learning (MIL) Given that biomarker labels are typically available only at the patient level, MIL frameworks with attention mechanisms aggregate tile-level features into slide-level predictions [52]. The attention weights identify morphological regions most predictive of biomarker status [52].
Data Augmentation and Stain Normalization To address variability in staining protocols and scanner systems, traditional approaches employ extensive data augmentation and stain normalization techniques like the Vahadane method [56].
Validation Frameworks
Robust validation is essential for both approaches:
Multi-Center Validation Models are evaluated on internal hold-out sets and external datasets from different institutions to assess generalizability across scanner types, staining protocols, and patient populations [53] [43]. For example, EAGLE was validated on samples from five national and international institutions [53].
Prospective Clinical Validation The most rigorous validation involves prospective "silent trials" where models are integrated into clinical workflows without impacting patient care. In one such trial, EAGLE achieved an AUC of 0.890 on prospective samples, demonstrating real-world clinical utility [53].
Benchmarking Platforms Standardized benchmarks like the EVA platform provide objective performance comparisons across models on diverse tasks including tile classification, nuclear segmentation, and slide-level diagnosis [54].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key research reagents and computational resources for biomarker prediction studies
| Resource Category | Specific Tools | Function/Purpose |
|---|---|---|
| Foundation Models | Virchow, PLUTO-4, TITAN, UNI | Pre-trained feature extractors for transfer learning [8] [43] [54] |
| Traditional Models | ResNet, DenseNet, Vision Transformers | Backbone architectures for custom model development [52] |
| Whole Slide Image Datasets | TCGA, In-house institutional archives | Training and validation data sources [53] [43] |
| Annotation Tools | VGG Image Annotator (VIA), Digital pathology platforms | Region-of-interest marking and label generation [56] |
| Stain Normalization | Vahadane method, Macenko method | Standardizing color distributions across slides [56] |
| Multiple Instance Learning Frameworks | Attention-MIL, TransMIL | Weakly supervised learning for slide-level predictions [52] |
| Biomarker Ground Truth | MSK-IMPACT, Idylla, NGS panels | Reference standards for model training and validation [53] |
The comparative analysis demonstrates distinct advantages and limitations for both foundation models and traditional transfer learning approaches in predicting genetic alterations from H&E morphology.
Foundation models consistently outperform traditional approaches across multiple biomarkers and cancer types, particularly in scenarios with limited training data, rare cancers, and cross-institutional generalization [8] [43]. Their large-scale pretraining on diverse histopathological data enables robust feature representations that transfer effectively to various downstream prediction tasks. The integration of multimodal capabilities further expands their utility for cross-modal retrieval and report generation [8].
Traditional transfer learning approaches remain valuable for targeted applications with sufficient training data and well-defined morphological correlates. Their simpler architecture requirements and established methodologies offer practical advantages for focused biomarker prediction tasks [52] [56].
For researchers and drug development professionals, selection between these approaches should consider available computational resources, dataset size and diversity, and specific clinical application requirements. Foundation models represent the advancing frontier of computational pathology, while traditional approaches provide accessible entry points for biomarker discovery initiatives.
Future directions include developing more efficient foundation model architectures, improving interpretability for clinical adoption, and establishing standardized benchmarks for objective performance assessment across the rapidly evolving landscape of computational pathology.
Computational pathology is undergoing a paradigm shift, moving from task-specific models trained via traditional transfer learning to versatile foundation models pretrained on massive datasets. Foundation models are a subclass of deep learning models trained on especially broad datasets and designed to be applicable to a range of different downstream tasks [11]. In computational pathology, these models capture comprehensive histomorphological patterns from millions of tissue regions, creating a versatile "visual vocabulary" that can be adapted to various clinical tasks with minimal additional training [8] [57].
Traditional transfer learning typically involves taking a model pretrained on a general dataset (e.g., ImageNet) and fine-tuning it on a specific pathology task with a limited labeled dataset. This approach has enabled many deep learning applications but requires extensive task-specific labeling and often struggles with generalizability [1]. In contrast, foundation models like TITAN (Transformer-based pathology Image and Text Alignment Network) are pretrained using self-supervised learning on hundreds of thousands of whole-slide images, learning robust feature representations without clinical labels [8]. This fundamental difference in training approach has significant implications for clinical workflow integration, particularly in diagnostic assistance, prognostication, and report generation.
Performance Comparison: Quantitative Analysis
Slide-Level Classification and Prognostication Performance
Table 1: Performance comparison (%) of foundation models versus traditional transfer learning on slide-level tasks
| Model Type | Model Name | Linear Probing (Avg) | Few-Shot Learning (Avg) | Zero-Shot Classification | Rare Cancer Retrieval |
|---|---|---|---|---|---|
| Foundation Model | TITAN (Proposed) | 85.4 | 80.7 | 76.2 | 84.9 |
| Foundation Model | TITANV (Vision-only) | 83.1 | 78.3 | N/A | 82.5 |
| Multimodal Slide Foundation Model | Previous State-of-the-Art | 79.8 | 72.1 | 65.3 | 75.6 |
| ROI Foundation Model | CONCH-based | 75.2 | 68.9 | N/A | 70.3 |
| Traditional Transfer Learning | Supervised Baseline | 72.5 | 60.4 | N/A | 65.8 |
The TITAN foundation model demonstrates superior performance across multiple machine learning settings, particularly in low-data regimes and specialized tasks like rare cancer retrieval [8]. Without any fine-tuning or requiring clinical labels, TITAN can extract general-purpose slide representations that outperform both region-of-interest (ROI) and other slide foundation models. The model's strong zero-shot classification capability (76.2%) is particularly notable, as it enables application to diagnostic scenarios without task-specific training data [8].
Cross-Modal Retrieval and Report Generation Performance
Table 2: Multimodal capabilities comparison across model architectures
| Model Type | Cross-Modal Retrieval (mAP) | Report Generation (BLEU Score) | Clinical Relevance Score | Diagnostic Accuracy from Generated Reports | |
|---|---|---|---|---|---|
| Multimodal Foundation Model | TITAN | 0.812 | 0.745 | 4.31/5 | 92.7% |
| Vision-Language Pretrained Model | Previous VLP | 0.723 | 0.682 | 3.95/5 | 88.3% |
| Text Summarization Model | T5-based | N/A | 0.698 | 4.02/5 | 89.1% |
| Image-to-Text Generation | CNN-RNN | N/A | 0.621 | 3.74/5 | 85.6% |
TITAN's multimodal capabilities, achieved through vision-language alignment with pathology reports and synthetic captions, enable impressive cross-modal retrieval and report generation performance [8]. In radiology, similar AI report generation systems have achieved RadCliQ-v1 scores of 1.46±0.03 on findings sections, outperforming other AI systems in clinical relevance metrics [58]. Quality assessments of AI-generated reports show significant promise, with radiologists rating summary quality at 4.86/5 and recommendation agreement at 4.94/5 [58].
Experimental Protocols and Methodologies
Foundation Model Pretraining Protocol
The TITAN model employs a three-stage pretraining strategy to create general-purpose slide representations [8]:
Stage 1: Vision-only Unimodal Pretraining
- Dataset: 335,645 whole-slide images (Mass-340K) across 20 organs
- ROI Processing: Non-overlapping patches of 512×512 pixels at 20× magnification
- Feature Extraction: 768-dimensional features for each patch using CONCHv1.5
- SSL Framework: iBOT (knowledge distillation with masked image modeling)
- View Generation: Random cropping of 2D feature grid (16×16 features covering 8,192×8,192 pixels)
- Augmentation: Vertical/horizontal flipping with posterization feature augmentation
Stage 2: Cross-Modal Alignment at ROI-Level
- Dataset: 423,122 synthetic fine-grained ROI captions generated using PathChat
- Alignment: Contrastive learning between ROI features and textual descriptions
Stage 3: Cross-Modal Alignment at WSI-Level
- Dataset: 182,862 medical reports paired with whole-slide images
- Objective: Vision-language pretraining for slide-report alignment
Foundation Model Pretraining Workflow
Transfer Learning Optimization Protocol
For traditional transfer learning approaches, rigorous hyperparameter optimization is essential. A comprehensive study evaluated three transfer learning techniques across multiple foundation models [11]:
Hyperparameter Search Strategy
- Algorithm: Bayesian optimization with 3-fold cross-validation
- Parameters: Learning rate (1e-7 to 0.1), weight decay (1e-4 to 0.5), beta1 (0.4-0.9), beta2 (0.9-0.999)
- Validation: Stratified holdout set (10% of dataset) with patient-level separation
- Optimizer: AdamW with optimized hyperparameters
Transfer Learning Techniques Evaluated
- End-to-End Training: All learnable parameters trained for five epochs
- Embedding-Only Training: Only embeddings and classification head trained for five epochs
- Embedding-First Training: Embedding-only training for one epoch, followed by four epochs of end-to-end training
The results demonstrated that end-to-end training generally outperformed other knowledge transfer paradigms, with low learning rates and high weight decays proving most effective [11].
Evaluation Methodology
Downstream Task Evaluation Foundation models were evaluated on diverse clinical tasks including:
- Linear Probing: Frozen features with trained linear classifier
- Few-Shot Learning: Limited labeled examples (typically 1-16 per class)
- Zero-Shot Classification: No task-specific training, using natural language descriptions
- Rare Cancer Retrieval: Slide-level similarity search for rare disease cases
- Cross-Modal Retrieval: Text-to-slide and slide-to-text retrieval
- Report Generation: Automated generation of pathology findings
Statistical Validation All experiments employed patient-level data splitting to prevent data leakage, with comprehensive cross-validation and external validation where possible [8] [11].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential research reagents and computational resources for foundation model development
| Resource Category | Specific Tool/Solution | Function/Purpose | Key Features |
|---|---|---|---|
| Foundation Models | TITAN | General-purpose slide representation learning | Multimodal (image + text), 336K WSIs pretraining, zero-shot capabilities |
| Foundation Models | UNI | Vision transformer for pathology | RGB-trained, adaptable to various downstream tasks |
| Foundation Models | CONCH | Regional feature embedding | Patch-level representation, 768-dimensional features |
| Feature Extraction | CONCHv1.5 | Patch-level feature extraction | Extended version of CONCH, processes 512×512 patches |
| Data Generation | PathChat | Synthetic caption generation | Multimodal generative AI copilot for pathology, creates fine-grained descriptions |
| Multimodal Alignment | iBOT Framework | Self-supervised learning | Knowledge distillation with masked image modeling |
| Transfer Learning | Optimized Hyperparameter Sets | Model adaptation | Bayesian-optimized parameters for specific tasks |
| Evaluation Framework | Retrospective Evaluation Framework | Model validation | Standardized assessment for clinical AI applications [59] |
Technical Implementation and Workflow Integration
Computational Pathology Workflow
Computational Pathology Analysis Pipeline
The technical workflow begins with tissue sample preparation, which can include traditional H&E staining or immunohistochemical stains for additional phenotypic information [1]. Slides are digitally scanned to create whole-slide images (WSIs), which are typically divided into smaller patches or tiles for analysis since the gigapixel-sized WSIs cannot be directly processed by conventional neural networks [1]. Modern digital scanners can capture images at up to 40× magnification with multiple z-stack levels, producing high-resolution images (commonly 100k × 100k pixels) with comprehensive color information [1].
Integration with Clinical Decision Support
Foundation models enable multiple integration pathways for clinical decision support:
Diagnostic Assistance
- Priority Sorting: AI algorithms can flag suspicious cases for pathologist review
- Paige Prostate Detect: FDA-cleared AI tool demonstrating 7.3% reduction in false negatives [60]
- TITAN Zero-Shot: Classification without task-specific training data [8]
Prognostication Support
- ArteraAI Prostate: Multimodal AI-powered tool combining clinical data and histological images for prognosticating 10-year risk of distant metastasis [1]
- CNN-derived risk scores: Providing additional prognostic value compared to TNM staging in gastric cancer [1]
Report Generation
- Automated Findings: Generation of diagnostic descriptions from WSI analysis
- Structured Reporting: Consistent report formatting and terminology
- Cross-Modal Retrieval: Finding similar cases based on textual queries
The integration of foundation models into computational pathology represents a significant advancement over traditional transfer learning approaches. The quantitative evidence demonstrates that foundation models like TITAN achieve superior performance across multiple clinical tasks, particularly in low-data regimes and specialized applications like rare disease diagnosis. Their multimodal capabilities enable novel applications in cross-modal retrieval and report generation that extend beyond the capabilities of traditional approaches.
For researchers and drug development professionals, foundation models offer a versatile foundation that can accelerate research and development workflows. The ability to perform zero-shot and few-shot learning reduces dependency on extensive labeled datasets, while the multimodal understanding enables more comprehensive tissue analysis. As these models continue to evolve, they are poised to become indispensable tools in the computational pathology toolkit, transforming clinical workflows through enhanced diagnostic assistance, improved prognostication, and automated report generation.
Navigating Implementation Challenges: Robustness, Bias, and Optimization Strategies
Combatting Domain Shift and Site-Specific Bias in Multi-Center Deployment
The deployment of artificial intelligence (AI) models in computational pathology faces a significant obstacle: domain shift and site-specific bias. These phenomena occur when models trained on data from one institution perform poorly on data from new institutions due to variations in staining protocols, scanning equipment, tissue processing methods, and other technical factors [61] [62] [63]. Such performance degradation poses serious challenges for clinical implementation, particularly in multi-center research and drug development contexts. Within computational pathology research, two predominant approaches have emerged to address these challenges: foundation models pretrained on massive, diverse datasets using self-supervised learning, and traditional transfer learning methods that adapt existing models to new domains. This guide provides a comprehensive comparison of these approaches, examining their experimental performance, methodological frameworks, and practical implications for researchers and drug development professionals.
The core of the problem lies in the fact that deep learning models can learn spurious correlations with site-specific artifacts rather than biologically relevant features. Studies have demonstrated that pathology foundation models (PFMs) can achieve alarmingly high accuracy (approximately 70-95%) in identifying the source institution of an image, indicating that institution-specific signatures are deeply embedded in the extracted features [61] [64]. This "feature contamination" undermines model reliability in real-world clinical settings where generalizability across diverse healthcare institutions is paramount [64].
Domain shift in medical imaging AI arises from both technical and clinical factors. Technical sources include:
- Scanner Variability: Differences in MRI field strength (1.5T vs. 3T), coil design, gradient performance, and vendor-specific reconstruction algorithms (GE, Siemens, Philips) create distinct image characteristics [63].
- Staining Protocols: Variations in hematoxylin and eosin (H&E) staining intensity, tissue preparation, and staining techniques across pathology laboratories [61] [62].
- Acquisition Parameters: Differences in slide scanning resolution, compression settings, and image formatting protocols [64].
- Environmental Factors: Hardware aging, calibration drift between maintenance cycles, and storage protocols [63].
Clinical Consequences of Domain Shift
The impact of domain shift extends beyond technical metrics to tangible clinical implications:
- Performance Degradation: Models may maintain 95% accuracy on their training domain but plummet to 70% or lower when deployed at new institutions [63].
- Feature Instability: Radiomic and pathomic features used for biomarker development show poor reproducibility across different scanner models and acquisition protocols [63].
- Regulatory Challenges: Regulatory bodies like the FDA require evidence of robust performance across diverse equipment and sites, creating significant hurdles for approval [63].
- Equity Concerns: Performance disparities across demographic groups may be exacerbated if certain populations are underrepresented in training data [61].
Foundation Models vs. Traditional Transfer Learning: Conceptual Frameworks
Pathology Foundation Models (PFMs)
Foundation models represent a paradigm shift in computational pathology. These models are pretrained on massive, diverse datasets using self-supervised learning (SSL) objectives, without requiring expensive manual annotations [8] [65]. The resulting models learn general-purpose feature representations that can be adapted to various downstream tasks with minimal fine-tuning. Notable examples include:
- UNI: ViT-large model trained on 100 million tiles from 100,000 slides using DINOv2 algorithm [65].
- Virchow: ViT-huge model trained on 2 billion tiles from nearly 1.5 million slides [65].
- TITAN: A multimodal whole-slide foundation model pretrained on 335,645 whole-slide images via visual self-supervised learning and vision-language alignment [8].
- Prov-GigaPath: Employed tile-level pretraining with DINOv2 followed by slide-level pretraining using masked autoencoder and LongNet [65].
Traditional Transfer Learning Approaches
Traditional transfer learning encompasses a range of techniques designed to adapt models to new domains:
- Domain Adaptation: Methods like Adversarial Domain Adaptation (ADA) aim to align feature distributions between source and target domains [62].
- Data Augmentation: Techniques including color normalization, stain augmentation, and style transfer [62] [66].
- Specialized Algorithms: Approaches like Adversarial fourIer-based Domain Adaptation (AIDA) which leverages frequency domain information to enhance model robustness [62].
The following workflow diagram illustrates how these approaches tackle domain shift:
Diagram: Domain Shift Challenges and Solution Approaches in Computational Pathology
Experimental Performance Comparison
Quantitative Benchmarking Across Models
Recent comprehensive benchmarking studies have evaluated the performance of various public pathology foundation models and traditional approaches across multiple clinical tasks [65]. The table below summarizes the performance characteristics of leading foundation models:
Table 1: Pathology Foundation Models Performance Characteristics
| Model | Parameters (M) | Training Data | Algorithm | Key Strengths | Domain Shift Resilience |
|---|---|---|---|---|---|
| CTransPath | 28 | TCGA, PAIP (16M tiles) | SRCL | Strong performance on retrieval and classification | Moderate [65] |
| Phikon | 86 | TCGA (43M tiles) | iBOT | Balanced performance across tasks | Moderate [65] |
| UNI | 303 | MGB (100M tiles) | DINOv2 | Excellent slide-level classification | High [65] |
| Virchow | 631 | MSKCC (2B tiles) | DINOv2 | State-of-the-art on diverse benchmarks | High [65] |
| Prov-GigaPath | 1135 | PHS (1.3B tiles) | DINOv2 + MAE | Strong genomic prediction | High [65] |
Performance on Domain Shift Tasks
Experimental evaluations demonstrate significant differences in how foundation models and traditional approaches handle domain shift. The following table compares performance metrics across multiple studies:
Table 2: Performance Comparison Under Domain Shift Conditions
| Method | Cancer Types | In-domain Accuracy | Out-of-domain Accuracy | Performance Drop | Reference |
|---|---|---|---|---|---|
| AIDA (Traditional) | Ovarian, Pleural, Bladder, Breast | 89.2% | 85.7% | 3.5% | [62] |
| Foundation Models (Avg.) | Multiple (17 tasks) | 91.8% | 88.3% | 3.5% | [65] |
| Stain Normalization | Lung, Kidney | 84.5% | 76.2% | 8.3% | [64] |
| Adversarial Training | Prostate MRI | 87.9% | 82.1% | 5.8% | [66] |
| Standard Supervised | Multiple | 92.4% | 74.6% | 17.8% | [61] |
Notably, the AIDA framework (Adversarial fourIer-based Domain Adaptation) demonstrates particularly strong performance in subtype classification tasks across four cancers, achieving superior classification results in the target domain compared to baseline methods, color augmentation, and standard adversarial domain adaptation [62].
Multi-Center Validation Performance
Recent research has highlighted the importance of rigorous multi-center validation. One study developing a multitask prediction model for postoperative outcomes demonstrated robust performance across three independent cohorts, with AUROCs for acute kidney injury prediction ranging from 0.789 to 0.863 across sites [67]. This underscores the potential of approaches specifically designed for generalizability.
Methodological Deep Dive: Experimental Protocols
Foundation Model Pretraining Protocol
The training methodology for foundation models like TITAN typically involves a multi-stage process [8]:
Vision-Only Unimodal Pretraining:
- Utilizes self-supervised learning (SSL) on large-scale WSI collections (e.g., 335,645 slides)
- Employs techniques like masked image modeling and knowledge distillation (iBOT framework)
- Processes non-overlapping patches of 512×512 pixels at 20× magnification
- Extracts 768-dimensional features for each patch using specialized encoders
Cross-Modal Alignment at ROI-Level:
- Aligns visual features with synthetic fine-grained morphological descriptions
- Uses generated captions from multimodal generative AI (423k ROI-caption pairs)
Cross-Modal Alignment at WSI-Level:
- Aligns entire slide representations with corresponding pathology reports
- Leverages 183k pairs of WSIs and clinical reports
This multi-stage approach enables the model to learn hierarchical representations that capture both local histological patterns and global slide-level context [8].
Traditional Domain Adaptation Protocol
The AIDA framework employs a specialized methodology to combat domain shift [62]:
Fourier Transform Enhancement:
- Incorporates an FFT-Enhancer module into the feature extractor
- Makes adversarial networks less sensitive to amplitude variations (color space changes)
- Increases attention to phase information (shape-based features)
Adversarial Training:
- Implements domain discrimination and feature extraction simultaneously
- Uses intermediate layer features for more effective domain adaptation
Multi-Center Validation:
- Evaluates on 1113 ovarian, 247 pleural, 422 bladder, and 482 breast cancer cases
- Employs comprehensive pathologist review to validate identified features
This approach specifically addresses the limitation that CNNs are more sensitive to amplitude spectrum variations while humans rely more on phase-related components for object recognition [62].
The following diagram illustrates the experimental workflow for evaluating domain shift resilience:
Diagram: Experimental Workflow for Domain Shift Resilience Evaluation
Successful research in combatting domain shift requires specific computational tools and datasets. The following table details key resources mentioned in recent literature:
Table 3: Essential Research Reagents for Domain Shift Studies
| Resource | Type | Key Features | Application in Domain Shift Research |
|---|---|---|---|
| TCGA Dataset | Pathology Images | 32,072 WSIs, 29 cancer types, 156 data centers | Benchmarking domain shift across centers [61] |
| AIDA Framework | Algorithm | Adversarial Fourier-based Domain Adaptation | Improving cross-domain classification [62] |
| TITAN Model | Foundation Model | Multimodal whole-slide foundation model | Zero-shot classification and report generation [8] |
| AdverIN | Domain Generalization | Monotonic adversarial intensity attacks | Medical image segmentation generalization [66] |
| DS-CP Framework | Uncertainty Quantification | Domain-shift-aware conformal prediction | Reliable uncertainty estimates under domain shift [68] |
| Stain Normalization | Preprocessing | Color distribution alignment | Reducing staining variations across sites [64] |
| Multi-center Validation Cohorts | Dataset | Independent patient populations from different sites | Rigorous generalizability testing [67] |
The comparison between foundation models and traditional transfer learning approaches for combatting domain shift reveals a complex landscape with distinct advantages for each paradigm. Foundation models excel through their ability to learn from massive, diverse datasets without manual annotation, capturing robust feature representations that generalize well across institutions. Their performance stems from scale, diverse pretraining data, and sophisticated self-supervised learning objectives [8] [65]. Conversely, traditional domain adaptation methods like AIDA offer targeted solutions that can be more parameter-efficient and specifically optimized for particular types of domain shift, such as those addressable through frequency domain manipulation [62].
The future of combatting domain shift in computational pathology likely lies in hybrid approaches that leverage the strengths of both paradigms. Promising directions include:
- Federated Foundation Models: Training large-scale models across institutions without sharing sensitive patient data [63].
- Synthetic Data Augmentation: Generating diverse training samples that simulate domain variations [66] [63].
- Domain Generalization Techniques: Moving beyond adaptation to specific known domains toward models that generalize to completely unseen domains [63].
- Multimodal Learning: Integrating pathology images with clinical reports and molecular data to learn more robust, biologically grounded representations [8].
For researchers and drug development professionals, the selection between foundation models and traditional transfer learning should be guided by specific use cases, available computational resources, and the diversity of intended deployment environments. Foundation models offer powerful off-the-shelf solutions for organizations with limited domain-specific data, while traditional approaches provide targeted optimization for specific domain shift challenges. As both paradigms continue to evolve, the gap between laboratory performance and real-world clinical utility will progressively narrow, ultimately accelerating the adoption of AI in pathology and drug development.
In computational pathology (CPath), the transition to digital whole-slide images (WSIs) has unlocked unprecedented potential for artificial intelligence (AI) to enhance diagnostic accuracy and efficiency [57]. These WSIs present a unique computational challenge due to their gigapixel resolutions and sparse diagnostic regions, which are typically analyzed using a weakly-supervised paradigm known as Multiple Instance Learning (MIL) [69]. A central debate in developing these AI models revolves around a critical strategic choice: what is the most effective pretraining strategy to ensure models generalize well to unseen data and clinical tasks? Is it the volume of data, or the diversity of that data?
This question sits at the heart of a broader thesis comparing two leading approaches: the emerging paradigm of foundation models—large models trained on vast, broad datasets—and traditional transfer learning, which often involves adapting existing models pretrained on more specific tasks [70] [69]. Recent systematic investigations reveal a compelling finding: models pretrained on strategically diverse, multi-task datasets consistently match or surpass the performance of models trained on larger, but less varied, data collections [69]. This guide provides an objective comparison of these approaches, presenting quantitative experimental data to help researchers and drug development professionals make informed decisions in their model development pipelines.
Core Concepts: Foundation Models vs. Traditional Transfer Learning
Foundation Models in Pathology
An AI foundation model is defined as a model trained on broad data at scale, generally using self-supervision, making it adaptable to a wide range of downstream tasks [70]. In CPath, these are often large-scale models designed to extract general-purpose slide-level representations that can transfer to challenging clinical tasks with little-to-no additional training (fine-tuning) [69] [57]. They aim to be a one-stop, reusable infrastructure for various applications.
Traditional Transfer Learning with MIL
In contrast, traditional transfer learning in CPath frequently employs Multiple Instance Learning (MIL). The MIL framework treats a whole slide image (WSI) as a "bag" containing thousands of individual image patches ("instances") [69]. A model learns to map the collection of patches to a slide-level diagnosis without needing patch-level annotations. In this context, traditional transfer learning involves taking a MIL model that was pretrained on a specific, supervised task (e.g., cancer subtyping on one organ) and adapting it to a new, different task (e.g., grading on another organ) [69].
Experimental Comparison: Diversity vs. Volume
Quantitative Performance Comparison
A systematic evaluation of 11 MIL models across 21 pretraining and target tasks provides robust data to compare the two strategic approaches. The study assessed performance when models were pretrained on single-organ datasets versus diverse, pancancer datasets, and then transferred to new, unseen tasks [69].
Table 1: Performance Comparison of Pretraining Strategies on Downstream CPath Tasks
| Pretraining Strategy | Pretraining Data Characteristic | Average Accuracy on Downstream Tasks | Key Strengths |
|---|---|---|---|
| Pancancer Pretraining | High Diversity (Multiple organs & task types) | Consistently Higher | Superior generalization across organs and task types; data-efficient [69]. |
| Single-Organ Pretraining | High Volume (Single organ) | Moderate | Good performance on tasks similar to the pretraining task [69]. |
| Training from Scratch | No Pretraining (No prior knowledge) | Lowest | Serves as a baseline; struggles in low-data regimes [69]. |
The core finding was that "pretrained MIL models consistently outperform MIL models trained with randomly initialized weights, even when pretrained on out-of-domain tasks." Furthermore, "models pretrained on pancancer tasks are data-efficient and generalize effectively across organs and task types," even outperforming some slide foundation models while using "substantially less pretraining data" [69].
Generalization Across Task Types
The advantage of diverse pretraining holds across different types of clinical challenges.
Table 2: Model Generalization Across Different CPath Task Types
| Target Task Type | Example | Performance of Pancancer-Pretrained Model | Performance of Single-Organ-Pretrained Model |
|---|---|---|---|
| Morphological Classification | Identifying basal cell carcinoma subtypes [71] | High Accuracy (e.g., >82% ACC) [71] [69] | Lower, more variable accuracy |
| Cancer Grading | Assessing tumor aggressiveness | Strong Generalization [69] | Limited generalization to different organs |
| Molecular Subtyping | Predicting biomarker status from histology | Effective Transfer [69] | Poor transfer if pretraining lacked molecular data |
Methodological Deep Dive: Experimental Protocols
Workflow for Evaluating Pretraining Strategies
The following diagram illustrates the core experimental protocol used to generate the comparative data, highlighting the comparison between leveraging a diverse pretrained model versus training from scratch.
The Multiple Instance Learning (MIL) Framework
The following diagram details the core MIL architecture, which is fundamental to many modern computational pathology models, whether used for traditional transfer learning or as a component in foundation models.
Key Experimental Protocol Details
The comparative findings are based on a rigorous experimental protocol [69]:
- Model Architectures: 11 different MIL models were evaluated, including ABMIL, CLAM, DSMIL, TransMIL, and simple baselines like MeanMIL and MaxMIL. This ensures findings are not architecture-specific.
- Pretraining Tasks: Models were pretrained on 21 distinct tasks, encompassing multiple organs (breast, lung, prostate, brain) and task types (cancer classification, grading, molecular subtyping).
- Transfer Evaluation: Pretrained models were then transferred to 19 downstream target tasks. Performance was evaluated using two methods:
- End-to-end fine-tuning: All model weights are updated on the new task.
- Frozen feature evaluation: The pretrained model's weights are frozen, and a simple classifier (like a K-Nearest Neighbors model) is used on the extracted slide-level features, testing the generality of the learned representations.
- Performance Metrics: Standard metrics including Accuracy and Area Under the Curve (AUC) were used for comparison against models trained from scratch with random weight initialization [71] [69].
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Reagents and Computational Tools for CPath Research
| Item | Function in Research | Example/Note |
|---|---|---|
| Whole Slide Images (WSIs) | The primary raw data; gigapixel digital scans of tissue sections [69]. | Sourced from hospital archives or public datasets; require careful annotation by pathologists [71]. |
| Multiple Instance Learning (MIL) Models | The core algorithmic framework for weakly-supervised slide-level classification [69]. | Examples: ABMIL, CLAM, TransMIL. Different aggregators introduce different inductive biases. |
| Pretrained Patch Encoders | Feature extraction backbones that convert image patches into numerical feature vectors [69]. | Often a CNN (e.g., ResNet) pretrained on natural images (ImageNet) or histopathology-specific datasets. |
| Pancancer Pretraining Datasets | Curated, diverse datasets used for strategic model pretraining. | Comprise WSIs from multiple cancer types and organs, enabling the learning of generalizable features [69]. |
| Computational Frameworks | Software libraries that standardize implementation and evaluation. | The "MIL-Lab" GitHub library provides a standardized resource for implementing MIL models and loading pretrained weights [69]. |
| High-Performance Computing (GPU Clusters) | Infrastructure for training and fine-tuning large models. | Essential for handling the computational load of processing gigapixel WSIs and large model architectures [69] [70]. |
The experimental evidence leads to a clear conclusion: in the pursuit of optimal generalization for computational pathology models, strategic emphasis on data diversity delivers superior returns compared to a narrow focus on data volume alone. Pancancer pretraining, which embodies this strategic diversity, produces models that are more data-efficient and robust when adapting to new organs, disease indications, and task types. While large-scale foundation models represent a powerful and evolving frontier, traditional transfer learning with MIL models—when pretrained on diverse, supervised tasks—offers a highly effective and often more data-efficient alternative. For researchers and drug developers building diagnostic and prognostic tools, prioritizing the curation of multi-faceted, heterogeneous datasets is a critical step toward creating AI models that truly generalize in the complex and varied real world of clinical practice.
The integration of artificial intelligence into computational pathology presents a critical trade-off between performance and practicality. On one hand, foundation models represent a transformative advance: large-scale models pretrained on massive, diverse datasets that can be adapted to numerous downstream tasks with minimal fine-tuning [57] [22]. Conversely, traditional transfer learning approaches, particularly those based on Multiple Instance Learning (MIL), offer a more established pathway that leverages existing architectures and smaller, targeted datasets [72] [73]. This comparison guide objectively evaluates both approaches through the critical lenses of computational overhead, energy requirements, data efficiency, and ultimate performance across key pathological tasks to inform researcher selection criteria.
Performance Comparison Tables
Table 1: Comparative Overview of Model Characteristics and Resource Demands
| Feature | Foundation Models (e.g., TITAN) | Traditional Transfer Learning (MIL) |
|---|---|---|
| Pretraining Data Scale | 335,645+ WSIs; 100M+ patches [8] [22] | Task-specific datasets; can leverage pancancer pretraining [72] |
| Architecture | Vision Transformer (ViT) for whole-slide encoding [8] | CNN-based (e.g., ResNet50) with MIL pooling [73] |
| Computational Load | Very High (Tens of thousands of GPU hours) [22] | Moderate (Fine-tuning requires significantly less compute) [72] [73] |
| Key Strength | Zero-shot learning, multimodal capabilities, high accuracy on rare tasks [8] | Efficient transfer across organs/tasks, strong performance with less data [72] |
| Major Limitation | Immense upfront compute and data requirements [8] [22] | May plateau below foundation model performance on complex tasks [72] |
Table 2: Quantitative Performance Comparison on Diagnostic Tasks
| Task / Metric | Foundation Model (Reported Performance) | Traditional Transfer Learning (Reported Performance) | Notes |
|---|---|---|---|
| Cancer Subtyping (AUC) | ~0.95 (Virchow, 16 cancer types) [22] | 0.98 Accuracy/F1 (ResNet50, Metastasis Detection) [73] | MIL excels in specific tasks; FMs lead in pan-cancer generalization. |
| Rare Cancer Retrieval | Outperforms other models (TITAN) [8] | Not specifically evaluated in search results | A key advantage for foundation models in low-data scenarios. |
| Data Efficiency | Strong few-shot and zero-shot performance [8] | Pretrained MIL models outperform from-scratch training [72] | Both benefit from pretraining, but FMs require far more data initially. |
| Cross-Modal Retrieval | Enabled (TITAN, CONCH) [8] [22] | Not a standard capability | Foundation models uniquely integrate image and text. |
Experimental Protocols and Methodologies
Foundation Model Pretraining and Adaptation
Large-scale foundation models like TITAN follow a complex, multi-stage pretraining process. The initial vision-only unimodal pretraining uses self-supervised learning on millions of histology image regions-of-interest (ROIs), often employing frameworks like iBOT for masked image modeling and knowledge distillation [8]. The input involves creating a 2D feature grid from patch features (e.g., 768-dimensional features from a patch encoder) extracted from gigapixel whole-slide images (WSIs). A critical step for handling computational load is the use of region cropping, where random crops of 16x16 features (covering 8,192x8,192 pixels) are sampled from the WSI feature grid for processing [8]. This is followed by cross-modal alignment, where the model is fine-tuned using synthetic captions and pathology reports to learn the association between visual patterns and textual descriptions [8]. For downstream task adaptation, linear probing (training only a new linear classifier on top of frozen features) or few-shot fine-tuning are common efficient protocols that leverage the model's rich pretrained representations without full end-to-end retraining [8].
Traditional Transfer Learning Workflows
In contrast, traditional transfer learning, such as the MIL approach used for metastatic breast cancer detection, follows a more direct protocol [73]. The process begins with annotation transfer, where spatial annotations from immunohistochemistry (IHC) whole-slide images (WSIs) are transferred to corresponding hematoxylin and eosin (H&E) slides to generate segmentation masks for training [73]. The H&E WSIs are then divided into smaller patches (e.g., 200x200 pixels) to manage computational load [73]. A pretrained CNN (e.g., ResNet50 with weights initialized from ImageNet) is used as a feature extractor for each patch [73]. The model is then fine-tuned on the target task, with studies showing that integrating external public datasets (like Camelyon16) during this phase significantly boosts performance, sometimes more than data augmentation alone [73]. Finally, patch-level predictions are aggregated to reconstruct a slide-level diagnosis and generate color-coded probability maps for pathologist interpretation [73].
Transfer Learning for Novel Modalities
Protocols also exist for adapting foundation models to new data modalities, demonstrating a hybrid approach. One study fine-tuned RGB-trained pathology foundation models (UNI, CONCH) on hyperspectral imaging (HSI) data [11]. The methodology involved input layer modification, replacing the model's original 3-channel input layer with one accepting 87 spectral channels, with careful weight initialization based on the spectral sensitivity of the original RGB channels [11]. Researchers then systematically compared fine-tuning paradigms: end-to-end training, embedding-only training, and embedding-first training [11]. A hyperparameter search found that for this cross-modal transfer, low learning rates and high weight decay yielded optimal performance, with end-to-end fine-tuning generally outperforming other methods [11].
Workflow Visualization
Diagram 1: Computational Workflow Comparison. This diagram contrasts the foundational model pathway, characterized by massive upfront pretraining and flexible adaptation, with the traditional transfer learning approach, which uses more moderate, task-specific fine-tuning.
The Scientist's Toolkit: Key Research Reagents and Materials
Table 3: Essential Computational and Data Resources for Pathology AI
| Resource / Solution | Function / Purpose | Relevance to Model Type |
|---|---|---|
| Whole-Slide Images (WSIs) | Digital representations of histology slides; the primary data input. | Essential for both paradigms. Foundation models require orders of magnitude more (100k+) [22]. |
| Patch Encoders (e.g., CONCH) | Extract meaningful feature representations from small image regions. | Core building block for foundation models like TITAN [8]. |
| Self-Supervised Learning (SSL) | Pretraining method that uses unlabeled data to learn general representations. | The cornerstone of foundation model pretraining [8] [22]. |
| Multiple Instance Learning (MIL) | A weakly supervised learning paradigm for whole-slide classification. | The backbone of many traditional transfer learning approaches [72] [73]. |
| Synthetic Captions/Data | Algorithmically generated text or data used for training. | Used by TITAN for vision-language alignment; growing role in overcoming data scarcity [8] [9]. |
| Vision Transformer (ViT) | Neural network architecture using self-attention for images. | The dominant architecture for new foundation models [8]. |
| Pre-annotated Public Datasets (e.g., Camelyon16) | Curated datasets with labels for specific tasks. | Crucial for training and benchmarking traditional MIL models; can boost their performance significantly [73]. |
The choice between foundation models and traditional transfer learning is not a matter of superiority but of strategic alignment with research constraints and objectives. Foundation models (TITAN, UNI, CONCH) offer unparalleled performance, flexibility, and advanced capabilities like zero-shot learning and multimodal reasoning, but demand immense computational resources and data [8] [22]. They are the optimal choice for institutions with significant infrastructure, ambitious pan-cancer projects, and exploratory research into new morphological biomarkers. Traditional transfer learning and MIL models provide a computationally efficient, data-parsimonious, and highly effective pathway for well-defined diagnostic tasks, such as metastasis detection, and are more readily deployable in resource-constrained settings [72] [73]. The emerging trend of using pancancer pretraining for MIL models further narrows the performance gap while maintaining a lower resource footprint [72]. Ultimately, the field is evolving toward a hybrid future, where the efficient adaptability of traditional methods complements the expansive capabilities of foundation models.
The adoption of artificial intelligence (AI) in computational pathology represents a paradigm shift in cancer diagnostics and research. However, the security and robustness of these systems against adversarial attacks and real-world noise have emerged as critical challenges. Within the broader debate comparing foundation models—large-scale models pre-trained on massive datasets—against traditional transfer learning approaches, security vulnerabilities provide a crucial axis for evaluation. Recent research reveals that the very architectures and training paradigms that enable high performance on clean benchmark datasets may exhibit dramatically different behaviors when confronted with malicious perturbations or naturally occurring image variations [74] [75]. This comparison guide examines the adversarial robustness of convolutional neural networks (CNNs), vision transformers (ViTs), and emerging pathology foundation models, providing experimental data to inform model selection for clinical and research applications.
Performance Comparison: Quantitative Robustness Across Model Architectures
Baseline Performance and Adversarial Susceptibility
Table 1: Performance Comparison Under Adversarial Attacks (Renal Cell Carcinoma Subtyping)
| Model Architecture | Baseline AUROC | AUROC under Low PGD Attack (ɛ=0.25e-3) | AUROC under Medium PGD Attack (ɛ=0.75e-3) | AUROC under High PGD Attack (ɛ=1.50e-3) | Relative Robustness |
|---|---|---|---|---|---|
| CNN (ResNet) | 0.960 [74] | 0.919 [74] | 0.749 [74] | 0.429 [74] | Low |
| CNN with Adversarial Training | 0.954 [74] | 0.951 [74] | 0.944 [74] | 0.932 [74] | Medium |
| Vision Transformer (ViT) | 0.958 [74] | 0.957 [74] | 0.955 [74] | 0.950 [74] | High |
| Foundation Model (Prov-GigaPath) | State-of-the-art on 25/26 tasks [76] | Limited published data on targeted attacks | Limited published data on targeted attacks | Limited published data on targeted attacks | Variable |
Table 2: Performance on Gastric Cancer Subtyping Under Attack
| Model Architecture | Baseline AUROC | AUROC under Low PGD Attack | AUROC under Medium PGD Attack | AUROC under High PGD Attack |
|---|---|---|---|---|
| CNN (ResNet) | 0.782 [74] | 0.380 [74] | 0.029 [74] | 0.000 [74] |
| CNN with Adversarial Training | 0.754 [74] | 0.731 [74] | 0.679 [74] | 0.595 [74] |
| Vision Transformer (ViT) | 0.768 [74] | 0.766 [74] | 0.763 [74] | 0.760 [74] |
The data reveals that while CNNs and ViTs perform comparably on non-attacked images, their robustness diverges significantly under adversarial pressure. ViTs maintain approximately 99% of their baseline performance even under high-strength attacks, whereas standard CNNs can degrade to near-random guessing [74]. This robustness advantage extends across both white-box and black-box attack scenarios, suggesting fundamental architectural benefits in ViTs' self-attention mechanisms that create more stable latent representations of clinically relevant morphological features [74].
Foundation Models: Scaling Advantages and Emerging Vulnerabilities
Foundation models represent a different approach, leveraging massive pre-training datasets and sophisticated architectures. Models like Prov-GigaPath (pretrained on 1.3 billion image tiles) and UNI (pretrained on 100 million images from 100,000+ slides) demonstrate state-of-the-art performance on numerous benchmarks [76] [32]. However, their robustness profile is complex. While their scale and diversity of training data potentially offer inherent regularization against some natural variations, studies reveal specific vulnerabilities.
Universal and Transferable Adversarial Perturbations (UTAP) represent a particularly concerning vulnerability for foundation models. These imperceptible noise patterns can collapse model embeddings across architectures and potentially transfer across institutions [75]. Furthermore, the representational spaces of many foundation models show troubling dependencies, with embeddings clustering more strongly by medical center or scanner type than by biological class—a phenomenon quantified by a Robustness Index (RI) where most models scored below 1 [75] [77].
Experimental Protocols: Methodologies for Assessing Robustness
Adversarial Attack Generation and Testing
Research into adversarial robustness employs standardized protocols to ensure comparable results across studies. For vulnerability assessment, Projected Gradient Descent (PGD) serves as the benchmark white-box attack, where adversaries have full model knowledge [74]. Attack strength is controlled primarily by the epsilon (ɛ) parameter, which constraints the perturbation magnitude. Studies typically evaluate multiple ɛ values (e.g., 0.25e-3, 0.75e-3, 1.50e-3) to measure performance degradation curves [74]. The detection threshold for human observers has been established at approximately ɛ=0.19 for CNNs and ɛ=0.13 for ViTs, ensuring attacks remain visually imperceptible during testing [74].
Additional attack types include:
- Fast Gradient Sign Method (FGSM): A single-step attack for rapid vulnerability assessment [74]
- AutoAttack (AA): An ensemble attack for reliable evaluation [74]
- Square Attack: A score-based black-box attack [74]
- Universal and Transferable Adversarial Perturbations (UTAP): Model-agnostic perturbations effective against foundation models [75]
Real-World Noise Simulation
Beyond malicious attacks, robustness against naturally occurring variations is equally critical. The imaging pipeline introduces multiple noise sources that can be approximated through synthetic corruption or measured through cross-site validation [75]:
- Staining Variation: Differences in H&E staining protocols across institutions
- Scanner Effects: Optical and illumination variability between scanner models
- Preparation Artifacts: Section thickness, dust, bubbles, and folding artifacts
- Processing Artifacts: Compression, color normalization, and format conversion issues
The robustness of foundation models is frequently assessed using a Robustness Index (RI), which quantifies whether model embeddings cluster more strongly by biological class versus medical center, with RI >1 indicating true biological robustness [75].
Table 3: Key Experimental Resources for Robustness Research
| Resource Category | Specific Examples | Research Application | Key Characteristics |
|---|---|---|---|
| Public Datasets | TCGA (The Cancer Genome Atlas) [76] [32] | Model training and validation | ~30,000 slides across 32 cancer types; widely used benchmark |
| Large-Scale Datasets | Prov-Path [76], Mass-100K [32] | Foundation model pre-training | 171,189 slides (Prov-Path); 100,000+ slides (Mass-100K); real-world diversity |
| Model Architectures | ResNet (CNN) [74], Vision Transformer (ViT) [74] | Architectural comparisons | CNNs: translation invariance; ViTs: global attention mechanisms |
| Adversarial Libraries | AutoAttack [74], Custom PGD implementations | Robustness evaluation | Standardized attack implementations for reproducible evaluation |
| Deployment Frameworks | WSInfer [78], QuPath [78] | Clinical integration | Open-source tools for model deployment and visualization |
| Benchmarks | Digital Pathology Benchmark [76], OncoTree Classification [32] | Performance validation | 26 tasks across subtyping and pathomics (Prov-GigaPath); 108 cancer types (UNI) |
The evidence demonstrates that security considerations should significantly influence model architecture decisions in computational pathology. For applications requiring maximum robustness against adversarial manipulation and real-world noise, Vision Transformers currently offer superior protection compared to equivalently trained CNNs, with minimal performance trade-offs [74]. While adversarial training strategies can partially harden CNNs, they require precise knowledge of potential attack vectors and slightly reduce baseline performance [74].
Foundation models present a more complex profile—their scale and diversity provide advantages in multi-task applications and data-efficient learning [76] [32], yet they introduce substantial computational demands and exhibit new vulnerabilities like UTAP attacks [75]. Critically, the representational spaces of many foundation models remain fragile, with embeddings overly sensitive to site-specific variations rather than biological signals [75] [77].
For clinical deployment, particularly in security-conscious environments, ViTs provide inherent robustness advantages. In research settings requiring flexible adaptation across multiple tasks, foundation models offer compelling capabilities, though their robustness should be rigorously validated across diverse real-world conditions before clinical integration. Future work must address the critical gap in standardized adversarial benchmarking for large-scale foundation models to enable comprehensive security assessments across the pathology AI landscape.
In the evolving landscape of modern medicine, artificial intelligence (AI) is emerging as a transformative force, reshaping the way healthcare is delivered by enabling early disease detection and personalized recommendations [79]. However, a critical limitation hinders its widespread adoption: the "black box" nature of many sophisticated AI models [79] [80]. This term refers to the inability to understand the internal decision-making process of an AI system, which can only be viewed in terms of its inputs and outputs [80]. In high-stakes domains like pathology and clinical medicine, this opacity is a major concern [80] [81]. Clinicians, who bear ultimate responsibility for patient care, require understanding of how a diagnosis is produced to validate the model’s logic and communicate findings effectively to patients [79] [81]. The lack of transparency can foster distrust, limit clinical acceptance, and raises ethical questions regarding accountability and potential bias [80] [81].
This challenge is acutely felt in computational pathology, where AI models are increasingly applied to complex whole-slide images (WSIs) [57]. A paradigm shift is underway, moving from traditional transfer learning, which adapts models pre-trained on general images to medical tasks, toward the development of specialized foundation models trained on massive datasets of histopathology images [65]. This guide provides a comparative analysis of these approaches, focusing on their performance, interpretability, and the methodologies used to benchmark them, with the aim of illuminating the path toward more transparent and trustworthy clinical AI.
Foundation Models vs. Traditional Transfer Learning: A Paradigm Shift
The fundamental difference between the two approaches lies in their training data and objectives. Traditional transfer learning typically involves taking a model like a Convolutional Neural Network (CNN) pre-trained on a large-scale natural image dataset (e.g., ImageNet) and fine-tuning it for a specific medical task [65]. While computationally efficient, this approach has a key limitation: the features the model learned from natural images (cats, cars, etc.) may not be optimally relevant or representative of the complex morphological patterns found in histopathology images [65].
In contrast, foundation models are trained from scratch using self-supervised learning (SSL) on very large, unlabeled datasets of histopathology images [65]. This method allows the model to learn fundamental, domain-specific representations of tissue structures, cellular arrangements, and other pathological features without the need for human-annotated labels [8] [65]. As noted in a comprehensive review, "It is becoming abundantly clear that using SSL to train image encoders on unlabeled pathology data is superior to relying on models pretrained on other domains such as natural images" [65].
Table 1: Core Conceptual Differences Between Approaches
| Feature | Traditional Transfer Learning | Pathology Foundation Models |
|---|---|---|
| Pre-training Data | Large datasets of natural images (e.g., ImageNet) | Massive datasets of unlabeled histopathology whole-slide images [65] |
| Pre-training Method | Supervised learning | Self-supervised learning (SSL) e.g., DINOv2, iBOT [8] [65] |
| Primary Advantage | Computational efficiency; requires less specialized data | Learns domain-specific features; superior performance and generalizability [65] |
| Key Challenge | Potential domain mismatch; less optimized for pathology | Requires immense computational resources and large-scale data curation [65] |
Performance Benchmarking: Experimental Data and Results
To objectively compare these approaches, the research community relies on rigorous benchmarking on a variety of clinically relevant tasks. Recent studies have established that foundation models consistently outperform models based on traditional transfer learning.
A key 2025 clinical benchmark of public self-supervised pathology foundation models systematically evaluated multiple models on a collection of datasets associated with cancer diagnoses and biomarkers [65]. The study concluded that SSL-trained pathology models hold immense potential and demonstrate clear superiority [65]. Furthermore, advanced foundation models like TITAN (Transformer-based pathology Image and Text Alignment Network) are designed to encode entire WSIs, simplifying slide-level clinical endpoint prediction and outperforming earlier patch-based models across tasks like cancer subtyping, biomarker prediction, and outcome prognosis [8].
The performance gap is evident in quantitative results. For instance, a hybrid ML framework integrating Explainable AI (XAI) strategies, which utilized ensemble models, reported an accuracy of 99.2% on tasks including predicting Diabetes, Heart Disease, and other conditions [79]. While not a foundation model per se, it highlights the potential of well-designed, interpretable systems. More directly, foundation models like UNI (a ViT-large model) and Virchow (a ViT-huge model), trained on 100 million and 2 billion pathology tiles respectively, have set new state-of-the-art performance on dozens of downstream tasks, including tissue classification and biomarker prediction [65].
Table 2: Benchmarking Performance of Select Pathology Foundation Models
| Model Name | Architecture | SSL Algorithm | Training Data Scale | Key Performance Highlights |
|---|---|---|---|---|
| UNI [65] | ViT-Large | DINOv2 | 100M tiles, 100k slides | Evaluated on 33 tasks; strong performance on classification and retrieval. |
| Virchow [65] | ViT-Huge | DINOv2 | 2B tiles, ~1.5M slides | State-of-the-art on tile-level and slide-level benchmarks. |
| Phikon [65] | ViT-Base | iBOT | 43M tiles, 6k slides | Assessed on 17 downstream tasks across 7 cancer indications. |
| TITAN [8] | ViT | iBOT & Vision-Language | 336k WSIs, 423k synthetic captions | Outperforms other slide foundation models in low-data regimes and zero-shot classification. |
| CTransPath [65] | Swin Transformer + CNN | SRCL (MoCo v3) | 16M tiles, 32k slides | Strong performance on WSI classification, mitosis detection, and segmentation. |
Key Experimental Protocols in Benchmarking
The methodology for benchmarking these models is critical for ensuring fair and reproducible comparisons. The following protocols are commonly employed:
Task Selection: Models are evaluated on a diverse set of clinically relevant tasks. These typically include:
- Slide-level classification: e.g., cancer subtyping, detection of metastases [65].
- Biomarker prediction: Predicting the status of genomic alterations (e.g., mutations) from histology images [8] [65].
- Prognosis: Predicting patient outcomes such as overall survival [65].
- Tile-level tasks: Image retrieval, patch classification, and segmentation [65].
Evaluation Metrics: Standard machine learning metrics are used to quantify performance. For classification tasks, these include Accuracy, Area Under the Receiver Operating Characteristic Curve (AUROC), Sensitivity, and Specificity [79] [65]. For retrieval tasks, metrics like recall are common [65].
Data Sourcing and Splitting: Benchmarks use datasets from multiple independent medical centers to test model generalizability. Data is rigorously split into training, validation, and test sets, often at the patient level to prevent data leakage and ensure a fair evaluation of the model's ability to generalize to new patients [65].
Fine-tuning Protocols: For foundation models, benchmarking often involves a process called linear probing, where only a simple linear classifier is trained on top of the frozen features extracted by the foundation model. This tests the quality of the learned representations themselves. This is compared to full fine-tuning, where more of the model's layers are updated for the specific task [8] [65].
Enhancing Interpretability: Methodologies for XAI
High performance alone is insufficient for clinical trust. To address the black box problem, Explainable AI (XAI) techniques are essential. These methods provide insights into which features of the input data most influenced the model's decision. Two of the most prominent model-agnostic techniques are:
SHAP (SHapley Additive exPlanations): A unified approach based on cooperative game theory that assigns each input feature an importance value for a particular prediction [79] [81]. This allows clinicians to see a quantitative breakdown of the factors leading to a diagnosis.
LIME (Local Interpretable Model-agnostic Explanations): Explains individual predictions by approximating the complex model locally with an interpretable model (e.g., a linear classifier) [79] [81]. It effectively creates a "local surrogate" model that is easier to understand.
In practice, these techniques can be integrated directly into a clinical workflow. For example, a hybrid ML-XAI framework for disease prediction combined models like Random Forests and XGBoost with SHAP and LIME to display the important features contributing to each prediction, thereby providing understandable explanations for interpretation of model outputs [79]. This enables clinical practitioners to make decisions through an understanding of AI-generated outputs.
The following diagram illustrates a generalized workflow for developing and explaining an AI model in computational pathology, integrating the concepts of foundation models and XAI.
Building, benchmarking, and explaining models in computational pathology requires a suite of tools and resources. The following table details key components essential for research in this field.
Table 3: Essential Research Reagents and Computational Tools
| Item / Resource | Type | Primary Function | Example Instances |
|---|---|---|---|
| Public Pathology Datasets | Data | Provides diverse, annotated data for training and benchmarking models. | TCGA (The Cancer Genome Atlas), PAIP [65] |
| SSL Algorithms | Software/Method | Enables pre-training of foundation models on unlabeled image data. | DINOv2, iBOT, Masked Autoencoders (MAE) [8] [65] |
| Model Architectures | Software/Model | The underlying neural network design for processing image data. | Vision Transformer (ViT), Swin Transformer, CNNs [8] [65] |
| XAI Libraries | Software Library | Generates post-hoc explanations for model predictions to enhance trust. | SHAP, LIME [79] [81] |
| Experiment Trackers | Software Platform | Manages the machine learning lifecycle, logging parameters and metrics for reproducibility. | MLflow, Weights & Biases (W&B) [82] |
| Whole-Slide Image (WSI) Encoders | Software/Model | Converts gigapixel WSIs into a sequence of lower-dimensional feature vectors for analysis. | CONCH [8] |
The journey toward fully trustworthy AI in clinical settings hinges on solving the black box problem. The evidence demonstrates that pathology-specific foundation models, trained via self-supervised learning on large-scale datasets, offer a dual advantage: they achieve superior predictive performance on a wide array of clinical tasks while also being more amenable to interpretation through modern XAI techniques [8] [65]. While traditional transfer learning provides a accessible starting point, the future of computational pathology is firmly rooted in the development and refinement of these powerful, transparent foundation models. As the field progresses, the continued integration of robust benchmarking, standardized explanatory methodologies, and collaborative input from clinicians, researchers, and regulators will be essential to translate this technological promise into safe, effective, and trusted patient care.
Benchmarks and Performance Analysis: Rigorous Comparative Evaluation
The emergence of foundation models represents a paradigm shift in computational pathology, moving away from the traditional transfer learning approach that dominated early research. Traditional transfer learning typically involved adapting models pre-trained on natural image datasets like ImageNet to histopathology data, a process limited by the significant domain gap between natural images and histopathological features. Foundation models, in contrast, are pre-trained directly on massive, diverse datasets of histopathology whole-slide images (WSIs) using self-supervised learning (SSL), capturing domain-specific morphological patterns without the need for extensive manual annotations [1] [83]. This report analyzes a comprehensive benchmark of 19 foundation models across 31 clinically relevant tasks to objectively evaluate their performance and utility for researchers and drug development professionals.
Performance Benchmarking Results
A large-scale independent evaluation benchmarked 19 histopathology foundation models on 13 patient cohorts comprising 6,818 patients and 9,528 slides from lung, colorectal, gastric, and breast cancers. The models were evaluated on 31 weakly supervised tasks related to biomarkers (19 tasks), morphological properties (5 tasks), and prognostic outcomes (7 tasks) [4].
Table 1: Top-Performing Foundation Models Across All Task Categories (Ranked by Mean AUROC)
| Foundation Model | Model Type | Morphology Tasks (Mean AUROC) | Biomarker Tasks (Mean AUROC) | Prognosis Tasks (Mean AUROC) | Overall Mean AUROC |
|---|---|---|---|---|---|
| CONCH | Vision-Language | 0.77 | 0.73 | 0.63 | 0.71 |
| Virchow2 | Vision-Only | 0.76 | 0.73 | 0.61 | 0.71 |
| Prov-GigaPath | Vision-Only | - | 0.72 | - | 0.69 |
| DinoSSLPath | Vision-Only | 0.76 | - | - | 0.69 |
| UNI | Vision-Only | - | - | - | 0.68 |
When averaged across all 31 tasks, CONCH and Virchow2 demonstrated the highest overall performance with AUROCs of 0.71, although their relative strengths varied across different task types [4]. CONCH, a vision-language model trained on 1.17 million image-caption pairs, performed on par with Virchow2, a vision-only model trained on a substantially larger set of 3.1 million WSIs, suggesting that architectural approach and data diversity can compensate for raw data volume [4].
Performance in Low-Data and Low-Prevalence Scenarios
A key advantage of foundation models is their potential utility in scenarios with limited labelled data, which is particularly relevant for rare molecular events or conditions. Benchmarking results revealed that performance advantages varied significantly in low-data settings [4].
Table 2: Performance in Data-Scarce Settings (Number of Tasks Where Model Ranked First)
| Foundation Model | Large Cohort (n=300) | Medium Cohort (n=150) | Small Cohort (n=75) |
|---|---|---|---|
| Virchow2 | 8 tasks | 6 tasks | 4 tasks |
| PRISM | 7 tasks | 9 tasks | 4 tasks |
| CONCH | - | - | 5 tasks |
In the largest sampled cohort (n=300 patients), Virchow2 demonstrated superior performance in 8 tasks, followed closely by PRISM with 7 tasks. With the medium-sized cohort (n=150), PRISM dominated by leading in 9 tasks, while Virchow2 followed with 6 tasks. The smallest cohort (n=75) showed more balanced results, with CONCH leading in 5 tasks, while PRISM and Virchow2 each led in 4 tasks [4]. These findings suggest that foundation model selection should be tailored to specific data availability contexts, with no single model dominating across all data regimes.
Experimental Protocols and Methodologies
Benchmarking Framework Design
The benchmarking study employed a standardized evaluation framework to ensure fair comparison across the 19 foundation models. The experimental protocol involved:
- Dataset Curation: 13 patient cohorts with 6,818 patients and 9,528 slides from lung, colorectal, gastric, and breast cancers, ensuring diversity in tissue sites, staining protocols, and scanner types [4].
- Task Selection: 31 clinically relevant tasks categorized into three domains: morphological properties (e.g., tissue structure, architecture), biomarkers (e.g., genetic mutations, protein expression), and prognostic outcomes (e.g., survival, treatment response) [4].
- Preprocessing Pipeline: WSIs were tessellated into small, non-overlapping patches, followed by image feature extraction using each foundation model. These features served as inputs for training downstream classification models [4].
- Evaluation Metrics: Primary evaluation used area under the receiver operating characteristic curve (AUROC), with additional validation using area under the precision-recall curve (AUPRC), balanced accuracy, and F1 scores [4].
Weakly Supervised Learning Approach
The benchmarking utilized weakly supervised multiple instance learning (MIL) to reflect real-world clinical scenarios where slide-level labels are more readily available than patch-level annotations. The methodology included:
- Feature Extraction: Each WSI was divided into tissue-containing patches, with features extracted using each foundation model without fine-tuning [4] [32].
- Aggregation Methods: Both transformer-based aggregation and attention-based multiple instance learning (ABMIL) were compared, with transformer-based approaches slightly outperforming ABMIL (average AUROC difference of 0.01) [4].
- Cross-Validation: To mitigate overfitting and ensure robust performance estimation, the evaluation employed external validation cohorts that were never part of any foundation model's training data, effectively addressing potential data leakage [4].
Weakly Supervised Learning Workflow: This diagram illustrates the standard pipeline for applying foundation models to whole-slide images using weakly supervised learning.
Visualization of Foundation Model Architectures
Foundation models in computational pathology employ diverse architectural strategies to process gigapixel whole-slide images and capture relevant morphological features at multiple scales.
Foundation Model Architectures in Pathology: This diagram categorizes the primary architectural approaches used by pathology foundation models and their shared learning methodologies.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents and Computational Resources for Pathology Foundation Model Research
| Resource Category | Specific Examples | Function in Research |
|---|---|---|
| Pathology Datasets | TCGA, CPTAC, Mass-100K, Mass-340K | Provide diverse, large-scale histopathology data for model training and validation across multiple tissue types and disease states [4] [32]. |
| Foundation Models | CONCH, Virchow2, UNI, Prov-GigaPath, TITAN, OpenMidnight | Serve as pre-trained feature extractors for downstream tasks, reducing need for task-specific model development from scratch [4] [8] [84]. |
| Evaluation Frameworks | eva, WSInfer, ABMIL, Multiple Instance Learning | Standardize performance assessment across tasks and models, enabling fair comparison and reproducibility [78] [84]. |
| Computational Resources | High-Performance GPUs (e.g., H100, AMD MI210), QuPath, Whole Slide Image Storage | Enable processing of gigapixel images, model training/inference, and visualization of results for pathologist interpretation [78] [84]. |
Discussion and Research Implications
Complementary Strengths and Ensemble Approaches
The benchmarking results revealed that foundation models trained on distinct cohorts often learn complementary features to predict the same label. Ensemble approaches combining multiple high-performing models consistently outperformed individual models, with a CONCH and Virchow2 ensemble outperforming individual models in 55% of tasks [4]. This suggests that rather than seeking a single superior model, researchers should consider hybrid approaches that leverage the complementary strengths of multiple foundation models, particularly for critical applications in drug development and diagnostic refinement.
Data Diversity Versus Scale
Contrary to conventional assumptions in deep learning, the benchmarking results challenge the notion that model performance scales monotonically with pretraining dataset size. While positive correlations (r = 0.29–0.74) were observed between downstream performance and pretraining dataset size, most were not statistically significant [4]. The success of CONCH, trained on 1.17 million image-caption pairs, compared to BiomedCLIP, trained on 15 million pairs, demonstrates that data diversity and quality may outweigh sheer volume in pathology foundation models [4]. This has important implications for resource-constrained research settings, suggesting that carefully curated, diverse datasets may be more valuable than massive, undifferentiated collections of histopathology images.
Emerging Capabilities and Clinical Translation
Foundation models are demonstrating capabilities that extend beyond traditional classification tasks, including resolution-agnostic tissue classification, few-shot class prototypes for slide classification, and cross-modal retrieval between histology slides and clinical reports [8] [32]. Models like TITAN can generate pathology reports and perform zero-shot classification, potentially reducing barriers to clinical adoption [8]. For drug development professionals, these capabilities offer new avenues for biomarker discovery, patient stratification, and treatment response prediction directly from routine H&E stains, potentially accelerating translational research pipelines.
In computational pathology, the development of artificial intelligence (AI) models has traditionally been constrained by the limited availability of large, annotated datasets. This challenge is particularly acute for rare diseases or molecular subtypes, where collecting sufficient training data is often impractical. Two dominant paradigms have emerged to address this issue: foundation models pretrained on vast, unlabeled datasets using self-supervised learning, and traditional transfer learning approaches, often based on Multiple Instance Learning (MIL), which leverage smaller, labeled datasets or models pretrained on natural images. This guide provides an objective comparison of their performance in data-scarce settings, synthesizing current benchmarking evidence to inform researchers and drug development professionals.
Independent, large-scale benchmarking studies reveal that while foundation models generally achieve superior performance, traditional approaches remain competitive, especially in specific scenarios. The following table summarizes the quantitative findings from recent comparative studies.
Table 1: Overall Performance Benchmarking of Foundation Models and Traditional Approaches
| Model Category | Representative Models (Top Performers) | Key Benchmark Findings | Performance in Low-Data Regimes |
|---|---|---|---|
| Pathology Foundation Models (Vision) | Virchow2, Prov-GigaPath, UNI [65] [85] | Achieved highest mean AUROC (0.706) across 19 TCGA tasks [85]; State-of-the-art in 25/26 clinical tasks [22] | Virchow2 dominated in settings with 300 and 150 patients; performance became more balanced with 75 patients [4] |
| Pathology Foundation Models (Vision-Language) | CONCH, PLIP [4] [65] | Highest overall performance (mean AUROC 0.71) in benchmarking of 31 tasks; excelled in morphology and prognosis [4] | Led in 5 out of 12 tasks with only 75 patients for training [4] |
| Traditional MIL & Transfer Learning | Pretrained MIL, ResNet50 (ImageNet) [72] [73] | Pretrained MIL models consistently outperformed models trained from scratch, even on different organs [72]; Achieved 0.98 accuracy in metastasis detection [73] | Effective for domain adaptation; outperforms slide foundation models with less pretraining data in some cases [72] |
Table 2: Performance on Specific Task Types (Mean AUROC)
| Model Type | Morphology Tasks | Biomarker Prediction | Prognosis Tasks | Notes |
|---|---|---|---|---|
| CONCH (Vision-Language FM) | 0.77 [4] | 0.73 [4] | 0.63 [4] | Trained on 1.17M image-text pairs [4] [65] |
| Virchow2 (Vision FM) | 0.76 [4] | 0.73 [4] | 0.61 [4] | Trained on 1.7B tiles from 3.1M WSIs [65] |
| Traditional MIL (from scratch) | Variable, generally lower | Variable, generally lower | Variable, generally lower | Performance highly dependent on task-specific data [72] |
Detailed Experimental Protocols and Methodologies
Benchmarking Foundation Models
Objective: To systematically evaluate the performance of pathology foundation models against traditional methods across diverse, clinically relevant tasks in data-scarce settings [4] [85].
Datasets:
- Foundation Model Pretraining Data: Models are pretrained on massive internal datasets (e.g., Mass-340K with 335,645 WSIs for TITAN [8], 3.1 million WSIs for Virchow2 [65]) comprising multiple organs, cancer types, and staining protocols.
- Downstream Evaluation Data: Benchmarking uses independent, external cohorts not seen during pretraining to ensure unbiased evaluation. For example, one study used 6,818 patients and 9,528 slides from lung, colorectal, gastric, and breast cancers [4]. Tasks include morphological classification, biomarker prediction (e.g., BRAF mutation), and survival prognosis [4] [85].
Workflow:
- Feature Extraction: A frozen foundation model is used as a feature extractor. Input whole-slide images (WSIs) are divided into smaller patches (e.g., 256x256 or 512x512 pixels), and each patch is converted into an embedding vector by the model [65] [85].
- Slide-Level Representation: The sets of patch embeddings for each WSI are aggregated into a single slide-level representation using a weakly supervised Multiple Instance Learning (MIL) aggregator, such as a transformer or an attention-based mechanism (ABMIL) [4] [85].
- Low-Data Protocol: To simulate data-scarce settings, downstream models are trained on randomly sampled subsets of the available labeled data (e.g., 75, 150, or 300 patients) while maintaining the original positive-to-negative case ratio [4].
- Evaluation: Model performance is assessed on held-out test sets using metrics like Area Under the Receiver Operating Characteristic curve (AUROC) and Area Under the Precision-Recall Curve (AUPRC) [4].
Diagram 1: Foundation Model Benchmarking Workflow
Evaluating Traditional Transfer Learning with MIL
Objective: To assess the transferability of traditional Multiple Instance Learning (MIL) models, particularly when pretrained on one organ or task and applied to another in a low-data setting [72] [73].
Datasets:
- Pretraining Data: MIL models are pretrained on specific, often public, pathology datasets (e.g., Camelyon16 for lymph node metastases [73]) or on pancancer datasets curated for a particular pretraining task [72].
- Target Data: The models are then evaluated on separate, target tasks which may involve different organs, cancer subtypes, or clinical endpoints (e.g., molecular subtype prediction) [72].
Workflow:
- Model Pretraining: A MIL model is trained from scratch on a source task with a sufficiently sized dataset. Alternatively, a CNN (e.g., ResNet50) pretrained on natural images (e.g., ImageNet) is used as a patch-level encoder [73].
- Transfer Learning: The pretrained model (or its encoder) is transferred to a new target task. In this step, the model weights can be either frozen, with only a new classification head trained, or fine-tuned entirely on the target data [72] [73].
- Low-Data Target Task: The key phase involves training or fine-tuning the model on the target dataset, which contains a very limited number of labeled slides (from tens to a few hundred) [72].
- Evaluation: Performance is measured on a held-out test set from the target task and compared against models trained from scratch and foundation model approaches [72].
Diagram 2: Traditional MIL Transfer Learning Workflow
Critical Analysis and Practical Considerations
Strengths and Limitations
Table 3: Analysis of Key Advantages and Challenges
| Aspect | Foundation Models | Traditional Transfer Learning/MIL |
|---|---|---|
| Data Efficiency | Excellent for zero-shot and few-shot learning once pretrained [8] [22]. | Requires a source task with adequate data for pretraining, but effective for subsequent low-data tasks [72]. |
| Computational Cost | Very high pretraining cost (thousands of GPU hours), but low cost for downstream adaptation [86] [22]. | Lower overall cost; fine-tuning is computationally cheaper than FM pretraining [86]. |
| Generalizability | Generally high, but can be confounded by site-specific bias (e.g., scanner, hospital) [86] [85]. | Good cross-organ generalization demonstrated, though may be task-dependent [72]. |
| Downstream Adaptation | Often limited to linear probing (training a shallow classifier on frozen features) due to instability of full fine-tuning [86]. | Allows for full fine-tuning of the model, providing greater flexibility for adaptation [72] [73]. |
| Robustness & Security | Shown to be vulnerable to universal adversarial perturbations, raising safety concerns [86]. | Less studied in this context, but may be less susceptible to certain attacks due to smaller capacity. |
The Scientist's Toolkit: Key Research Reagents and Materials
Table 4: Essential Resources for Computational Pathology Research
| Resource | Function in Research | Examples & Notes |
|---|---|---|
| Public Whole-Slide Image Repositories | Provide data for model pretraining, benchmarking, and low-data target tasks. | The Cancer Genome Atlas (TCGA), Camelyon16 [73] [65]. Essential for ensuring diversity and preventing data leakage in evaluations [85]. |
| Pretrained Model Weights | Enable researchers to leverage large-scale pretraining without the prohibitive cost. | Publicly released weights for models like CTransPath, Phikon, UNI, and Virchow [65]. |
| Multiple Instance Learning (MIL) Aggregators | Combine patch-level features into a slide-level prediction for both FM and traditional workflows. | Attention-based MIL (ABMIL) and Transformer aggregators are common choices [4] [85]. |
| Computational Hardware (GPUs) | Accelerate training and inference on gigapixel WSIs. | High-end GPUs (e.g., NVIDIA H100, A100) are needed for FM pretraining; less powerful cards suffice for fine-tuning and inference [86] [22]. |
| Standardized Benchmarking Pipelines | Allow for fair and reproducible comparison of different models across the same tasks. | Initiatives like the one from [65] provide automated pipelines for external validation. |
The evidence indicates that pathology foundation models like CONCH and Virchow2 currently set the benchmark for performance in data-scarce settings, offering robust off-the-shelf feature representations for diverse downstream tasks [4] [85]. However, traditional pretrained MIL models remain a potent and computationally efficient alternative, demonstrating remarkable transferability across organs and often outperforming slide foundation models when pretraining data is limited [72].
A promising future direction is model fusion, where ensembles combining top foundation models (e.g., CONCH and Virchow2) have been shown to outperform individual models in over 55% of tasks by leveraging their complementary strengths [4]. The field is also advancing towards more multimodal foundation models that integrate histology with pathology reports, genomic data, and other clinical information, further enhancing their utility in precision medicine and drug development [8] [22].
The field of computational pathology is undergoing a fundamental transformation, moving from traditional task-specific models toward more flexible foundation models. This evolution is characterized by a critical architectural choice: vision-only models that process histology images alone versus vision-language models (VLMs) that jointly understand images and textual information. Within the context of precision oncology, this distinction defines a new research paradigm where the integration of multimodal data determines a model's clinical utility, generalizability, and adaptability to diverse diagnostic scenarios.
Traditional computational pathology has relied predominantly on task-specific models, which require developing independent algorithms for each distinct clinical task—whether cancer classification, grading, or biomarker prediction. This approach depends heavily on large-scale annotated datasets, resulting in high costs, prolonged development cycles, and poor cross-task adaptability [87]. Foundation models, pretrained on vast amounts of data, promise to overcome these limitations. The emergence of pathology foundation models marks a pivotal shift toward "general intelligence" in computational pathology, enabling multi-task transfer with minimal or even zero annotated data, significantly enhancing clinical utility and generalizability [87].
This review systematically compares the capabilities of vision-language and vision-only foundation models within computational pathology, providing objective performance data, detailed experimental methodologies, and practical resources to guide researchers and drug development professionals in selecting appropriate architectures for precision oncology applications.
Model Architectures and Fundamental Differences
Vision-Only Foundation Models
Vision-only models in computational pathology are designed to extract critical visual features from whole slide images (WSIs) without integrating textual information. These models typically employ self-supervised learning techniques such as masked image modeling and knowledge distillation to learn powerful visual representations from unlabeled histopathology data [8]. Architecturally, they often utilize Vision Transformers (ViTs) that process sequences of patch embeddings extracted from gigapixel WSIs. Representative examples include GigaPath, UNI, and Virchow, which have achieved performance surpassing conventional approaches across various cancer types [87].
A key innovation in vision-only models is their approach to handling the computational challenge of processing extremely high-resolution WSIs. Models like TITAN (Transformer-based pathology Image and Text Alignment Network) construct input embedding spaces by dividing each WSI into non-overlapping patches, followed by extraction of patch features using specialized encoders [8]. These patch features are spatially arranged in a two-dimensional grid replicating their positions within the tissue, preserving spatial context crucial for pathological assessment.
Vision-Language Foundation Models
Vision-language models represent a more integrative approach, jointly processing visual information from histology images and textual data from pathology reports or synthetic captions. These models create a shared representation space where visual and linguistic concepts are aligned, enabling cross-modal understanding and retrieval. Notable examples include PLIP, CONCH, and PathChat, which leverage natural language annotations to comprehend image semantics and perform various downstream tasks [87].
The architectural paradigm for VLMs often involves multiple encoders—one for each modality—that fuse embeddings together to create a unified representation [88]. Decoders then use this shared latent space to generate outputs in the desired modality. For instance, Qwen2.5-Omni employs a novel "Thinker-Talker" architecture where the "Thinker" handles text generation and the "Talker" produces natural speech responses [88]. This architectural flexibility enables capabilities such as cross-modal retrieval, diagnostic report generation, and educational assistance in pathology.
Table 1: Comparison of Model Architectures and Training Approaches
| Feature | Vision-Only Models | Vision-Language Models |
|---|---|---|
| Primary Input | Whole slide images (WSIs) | WSIs + text (reports, captions) |
| Core Architecture | Vision Transformers (ViTs) | Multi-encoder frameworks with fusion mechanisms |
| Training Approach | Self-supervised learning (SSL) | Multimodal pretraining with alignment |
| Representative Examples | GigaPath, UNI, Virchow, TITAN-V | PLIP, CONCH, PathChat, TITAN |
| Key Innovation | Handling gigapixel WSIs via patch embedding | Cross-modal alignment of visual and textual concepts |
Performance Benchmarking and Comparative Analysis
Quantitative Performance Across Diagnostic Tasks
Recent comprehensive benchmarking studies reveal distinct performance patterns between vision-language and vision-only models. A systematic evaluation of 31 AI foundation models for computational pathology, including general vision models (VM), general vision-language models (VLM), pathology-specific vision models (Path-VM), and pathology-specific vision-language models (Path-VLM) across 41 tasks demonstrated that Virchow2, a pathology foundation model, delivered the highest performance across TCGA, CPTAC, and external tasks [21]. The study also showed that Path-VM outperformed both Path-VLM and VM, securing top rankings across tasks despite lacking a statistically significant edge over vision models [21].
In specialized evaluations focusing on diagnostic accuracy, Qwen2-VL-72B-Instruct achieved superior performance with an average score of 63.97% on the PathMMU dataset, outperforming other models across all subsets including PubMed, SocialPath, and EduContent [89]. This extensive evaluation of over 60 state-of-the-art VLMs revealed that model size alone does not guarantee superior performance, as effective domain alignment and domain-specific training are critical factors [89].
For zero-shot diagnostic pathology, studies investigating VLMs like Quilt-Net, Quilt-LLaVA, and CONCH on digestive pathology datasets comprising 3,507 WSIs found that prompt engineering significantly impacts model performance, with the CONCH model achieving the highest accuracy when provided with precise anatomical references [90]. This highlights the importance of anatomical context in histopathological image analysis, as performance consistently degraded when reducing anatomical precision.
Specialized Capabilities and Clinical Applications
Beyond overall accuracy, vision-language models demonstrate unique advantages in specialized capabilities crucial for clinical applications:
Cross-Modal Retrieval and Report Generation: Vision-language models excel at connecting visual patterns with textual descriptions, enabling content-based image retrieval using textual queries and automatic generation of pathology reports from whole slide images. TITAN, for instance, can generate pathology reports that generalize to resource-limited clinical scenarios such as rare disease retrieval and cancer prognosis [8].
Zero-Shot and Few-Shot Learning: The semantic alignment between visual and textual representations allows VLMs to recognize novel pathological findings without task-specific training. This is particularly valuable for rare diseases with limited training data. Studies show that VLMs can achieve competitive performance in zero-shot settings when properly instructed with domain-appropriate prompts [90].
Robustness and Generalization: Comprehensive evaluations reveal concerning limitations in both architectural approaches. A systematic assessment of ten leading pathology foundation models across multiple institutions found that only Virchow2 achieved a Robustness Index (RI) >1.2, indicating that biological structure dominated site-specific bias, whereas all others had RI ≈1, meaning their embeddings grouped primarily by hospital or scanner rather than by cancer type [86].
Table 2: Performance Comparison Across Specialized Tasks
| Task Type | Vision-Only Models | Vision-Language Models | Key Findings |
|---|---|---|---|
| Zero-Shot Classification | Limited to visual similarity | Enabled through semantic alignment | CONCH achieves highest accuracy with anatomical prompts [90] |
| Cross-Modal Retrieval | Not applicable | Core capability | TITAN enables slide-text retrieval without fine-tuning [8] |
| Rare Cancer Identification | Moderate performance (21-68% F1) [86] | Enhanced through language guidance | Enables retrieval of rare cases via textual descriptions |
| Multi-Organ Generalization | Significant performance variability across organs | More consistent performance through semantic regularization | Organ-level F1 scores: kidneys 68% vs. lungs 21% for vision-only [86] |
| Report Generation | Not applicable | Core capability | TITAN generates clinically relevant pathology reports [8] |
Experimental Protocols and Methodologies
Benchmarking Framework Design
Comprehensive evaluations of pathology foundation models require carefully designed experimental protocols. The benchmarking study assessing 31 models across 41 tasks utilized datasets from TCGA, CPTAC, external sources, and out-of-domain collections to ensure rigorous assessment [21]. Performance was measured using multiple metrics including accuracy, F1 scores, and retrieval precision, with special attention to cross-institutional generalization through the Robustness Index (RI) that quantifies whether model embeddings cluster more strongly by biological class or by medical center [86].
For VLM evaluations, the PathVLM-Eval framework employed the PathMMU dataset, which includes subsets such as PubMed, SocialPath, and EduContent featuring diverse formats including multiple-choice questions designed to aid pathologists in diagnostic reasoning [89]. Utilizing VLMEvalKit, an open-source evaluation framework, researchers brought publicly available pathology datasets under a single evaluation umbrella, ensuring unbiased and contamination-free assessments of model performance [89].
Training and Fine-Tuning Protocols
The training methodologies for vision-language and vision-only models differ significantly in their approach to data utilization and optimization:
TITAN's Three-Stage Pretraining: This approach exemplifies the sophisticated training required for effective VLMs in pathology. Stage 1 involves vision-only unimodal pretraining on ROI crops using iBOT framework for knowledge distillation. Stage 2 performs cross-modal alignment of generated morphological descriptions at ROI-level (423k pairs of 8k×8k ROIs and captions). Stage 3 conducts cross-modal alignment at WSI-level (183k pairs of WSIs and clinical reports) [8].
Prompt Engineering for Zero-Shot Evaluation: Studies investigating zero-shot diagnostic pathology developed a comprehensive prompt engineering framework that systematically varies domain specificity, anatomical precision, instructional framing, and output constraints [90]. This approach demonstrates that carefully designed prompts significantly enhance VLM performance, with the CONCH model achieving highest accuracy when provided with precise anatomical references.
Linear Probing vs. Full Fine-Tuning: Interestingly, in computational pathology, foundation model downstream use is overwhelmingly limited to linear probing—training a shallow linear classifier on frozen embeddings rather than fine-tuning the model itself [86]. This dependency arises because most pathology FMs are too large, memory-intensive, and unstable to fine-tune on moderate-sized clinical datasets, with full fine-tuning frequently degrading accuracy relative to linear probing due to overfitting and catastrophic forgetting.
Table 3: Key Research Reagents and Computational Resources for Pathology Foundation Model Development
| Resource Category | Specific Examples | Function and Application |
|---|---|---|
| Pathology Datasets | Mass-340K (335,645 WSIs) [8], TCGA, CPTAC | Pretraining and benchmarking foundation models across multiple organs and cancer types |
| Patch Encoders | CONCHv1.5 [8], UNI, Virchow | Extracting visual features from histology image patches for slide-level representation learning |
| Evaluation Frameworks | VLMEvalKit [89], PathVLM-Eval [89] | Standardized assessment of model performance across diverse pathology tasks |
| Benchmark Datasets | PathMMU (PubMed, SocialPath, EduContent) [89] | Specialized evaluation of VLM capabilities in histopathology image understanding |
| Synthetic Data Tools | PathChat [8] | Generating fine-grained ROI captions for vision-language alignment (423k pairs in TITAN) |
| Robustness Metrics | Robustness Index (RI) [86] | Quantifying whether embeddings cluster by biology vs. site-specific bias |
Critical Challenges and Limitations
Technical and Methodological Constraints
Both vision-language and vision-only models face significant technical challenges in clinical deployment. Geometric fragility remains a concern, with studies showing that transformer-based architectures lack inherent rotational inductive bias, requiring explicit rotation augmentation during training to achieve acceptable invariance [86]. Resource burden presents another barrier, as foundation models can consume up to 35× more energy than task-specific models, raising sustainability concerns [86].
Perhaps most critically, security vulnerabilities threaten clinical reliability. Research has demonstrated that universal and transferable adversarial perturbations (UTAP)—imperceptible noise patterns—can collapse FM embeddings across architectures, degrading accuracy from ≈97% to ≈12% on attacked models [86]. These vulnerabilities have real-world analogues in routine pathology workflow variations, including differences in H&E staining, scanner optics, compression artifacts, and slide preparation imperfections.
Domain-Specific Implementation Barriers
The complexity of human tissue morphology presents unique challenges for both architectural approaches. As noted in critical assessments, "a child learns to recognize dogs by age two and breeds by seven; a pathologist requires more than twelve years of education to distinguish cancer subtypes based on tissue morphology" [86]. This semantic complexity exceeds what current foundation models can capture, particularly when using self-supervision strategies developed for natural images.
Furthermore, the "myth of the universal model" conflicts with the No Free Lunch theorem, which states that no single model excels across all problems [86]. Benchmarks reveal wide organ-dependent performance swings, with vision-only models achieving 68% F1 scores for kidneys but only 21% for lungs [86], underscoring the limits of universal architectures for heterogeneous pathology applications.
The comparative analysis of vision-language versus vision-only models in computational pathology reveals a complex landscape where architectural advantages are highly context-dependent. Vision-language models demonstrate superior capabilities in zero-shot learning, cross-modal retrieval, and report generation, leveraging semantic alignment to handle diverse tasks with minimal fine-tuning. Conversely, vision-only models maintain advantages in computational efficiency and focused visual representation learning, particularly when task requirements are well-defined and data volumes substantial.
The emerging "Perception-to-Cognition" framework suggests that future advancements will require addressing deficits at both perceptual levels (fine-grained visual representation) and cognitive levels (multi-step reasoning) [91]. Next-generation models will likely incorporate more sophisticated observe-think-verify reasoning loops that dynamically re-examine visual evidence to validate or refine reasoning paths [91]. Additionally, addressing the fundamental challenges of biological complexity, geometric fragility, and site-specific bias will require domain-specific architectural innovations rather than direct transfers from general computer vision.
For researchers and drug development professionals, selection between vision-language and vision-only approaches should be guided by specific use case requirements: VLMs offer greater flexibility for exploratory research and educational applications, while vision-only models may provide more efficient solutions for focused diagnostic tasks with established visual criteria. As the field evolves, the integration of multimodal data—including genomic information—promises to further enhance the clinical utility of both architectural paradigms in precision oncology.
The field of computational pathology is undergoing a transformative shift from traditional transfer learning approaches to the use of foundation models pretrained on massive histopathology datasets. While transfer learning adapts models trained on natural images (e.g., ImageNet) to medical tasks, pathology foundation models are specifically pretrained on millions of histopathology images using self-supervised learning, capturing rich morphological patterns directly relevant to diagnostic applications [8] [4]. This specialized pretraining enables more robust performance across diverse clinical tasks, including cancer subtyping, biomarker prediction, and outcome prognosis [8].
However, individual foundation models exhibit distinct strengths and limitations based on their architectural designs, pretraining datasets, and learning objectives. No single foundation model consistently outperforms all others across every clinical scenario [4]. This limitation has catalyzed the emergence of ensemble strategies that strategically combine multiple foundation models to leverage their complementary strengths. By integrating predictions from diverse models, ensemble approaches achieve more accurate and reliable performance than any single model alone, particularly for challenging diagnostic tasks with significant clinical implications [92] [93].
Quantitative Comparison of Pathology Foundation Models
Performance Benchmarking Across Clinical Tasks
Recent comprehensive benchmarking studies have evaluated numerous pathology foundation models across clinically relevant tasks. One large-scale analysis assessed 19 foundation models on 31 weakly supervised downstream prediction tasks related to morphology, biomarkers, and prognostication using 6,818 patients and 9,528 slides [4]. The results demonstrated that while certain models consistently achieve strong performance, none dominates across all scenarios.
Table 1: Foundation Model Performance Across Task Types (AUROC)
| Foundation Model | Morphology Tasks (n=5) | Biomarker Tasks (n=19) | Prognosis Tasks (n=7) | Overall Average |
|---|---|---|---|---|
| CONCH (Vision-Language) | 0.77 | 0.73 | 0.63 | 0.71 |
| Virchow2 (Vision-Only) | 0.76 | 0.73 | 0.61 | 0.71 |
| Prov-GigaPath | 0.69 | 0.72 | 0.60 | 0.69 |
| DinoSSLPath | 0.76 | 0.68 | 0.60 | 0.69 |
| UNI | 0.68 | 0.68 | 0.61 | 0.68 |
The benchmarking revealed that CONCH, a vision-language model trained on 1.17 million image-caption pairs, and Virchow2, a vision-only model trained on 3.1 million whole-slide images, achieved equivalent overall performance [4]. Each excels in different contexts: CONCH demonstrates advantages in morphology-related tasks and overall metrics, while Virchow2 shows particular strength in biomarker prediction. This complementary performance profile makes them ideal candidates for ensemble integration.
Ensemble Performance Gains
Empirical studies consistently demonstrate that ensembles of foundation models achieve superior performance compared to individual models. Research on atypical mitosis classification shows that ensembles of multiple pathology foundation models can improve balanced accuracy by approximately 5% over the best-performing single model [92].
Table 2: Ensemble Model Performance for Atypical Mitosis Classification
| Model Type | Specific Models | Balanced Accuracy |
|---|---|---|
| Single Model | UNI | 85.46% |
| Single Model | Virchow | 86.04% |
| Single Model | Virchow2 | 87.59% |
| Ensemble Model | UNI + Virchow + Virchow2 | 93.57% |
Similarly, ensembles for central nervous system tumor diagnosis achieve exceptional performance, with the PICTURE system (Pathology Image Characterization Tool with Uncertainty-aware Rapid Evaluations) accurately distinguishing between glioblastoma and primary central nervous system lymphoma with an area under the receiver operating characteristic curve (AUROC) of 0.989, validated across five independent cohorts (AUROC = 0.924-0.996) [93].
Experimental Protocols for Ensemble Construction
Model Selection and Diversity Optimization
The foundation of effective ensemble construction lies in selecting models with diverse architectural characteristics and pretraining histories. Research indicates that models pretrained on distinct cohorts learn complementary features to predict the same label [4]. Optimal ensemble performance requires integrating models with varied inductive biases, which can be identified through their performance profiles across different task types.
For mitosis classification, researchers selected UNI, Virchow, and Virchow2 based on their complementary attention patterns over cellular structures, resulting from being pretrained on different collections of human histopathology images acquired under varying conditions (institutions, staining protocols, scanners) [92]. This diversity in training backgrounds enables the models to capture different morphological aspects of mitotic figures.
Weighted Ensemble Framework with Balanced Accuracy Optimization
A sophisticated weighted ensemble framework maximizes diagnostic accuracy while addressing class imbalance issues common in medical datasets. The approach involves learning optimal nonnegative weights for each base model's predictions on a validation set. Rather than maximizing overall accuracy (which may sacrifice performance on minority classes), the objective function directly maximizes balanced accuracy [92]:
For N base models producing probability vectors ( Pi(x) = [pi^{(1)}(x), \dots, pi^{(C)}(x)] ) over C classes, the ensemble learns weights ( wi ) by solving:
[ \mathbf{w}^* = \arg\max{\mathbf{w}} \frac{1}{C} \sum{c=1}^{C} \frac{1}{|D{\mathrm{val}}^c|} \times \sum{(x,y) \in D{\mathrm{val}}^c} \mathbf{1}\left[y = \arg\max{c'}\sum{i=1}^{N} wi p_i^{(c')}(x)\right] ]
subject to ( wi \geq 0, \sum{i=1}^{N} w_i = 1 )
Here, ( D_{\mathrm{val}}^c ) represents validation samples with label c. This formulation equally weights the accuracy of each class, mitigating performance degradation on minority classes [92].
Uncertainty Quantification and Out-of-Distribution Detection
Advanced ensemble systems incorporate epistemic uncertainty quantification to identify atypical pathology manifestations and enhance generalizability. The PICTURE framework employs Bayesian inference, deep ensemble, and normalizing flow techniques to account for uncertainties in predictions and training set labels [93]. This uncertainty-aware approach enables the system to correctly flag previously unseen central nervous system cancer types and normal tissues not represented in the training dataset, preventing overconfident and potentially misleading predictions.
Implementation Workflows and Methodologies
End-to-End Ensemble Construction Pipeline
The process of building effective ensembles for computational pathology follows a systematic workflow that integrates multiple foundation models with complementary strengths.
Parameter-Efficient Fine-Tuning with LoRA
Ensemble approaches often employ parameter-efficient fine-tuning techniques to adapt foundation models to specific diagnostic tasks without complete retraining. Low-Rank Adaptation (LoRA) introduces trainable rank decomposition matrices into transformer architectures while keeping original weights frozen [92]. For foundation models with query (Q) and value (V) projection matrices in multi-head self-attention modules, LoRA introduces low-rank matrices ( AQ \in \mathbb{R}^{d\times r} ) and ( BQ \in \mathbb{R}^{r\times k} ), so the update to the frozen query weight ( W_Q \in \mathbb{R}^{d\times k} ) is factorized as:
[ \Delta WQ = AQ BQ, \quad WQ = W0 + \Delta WQ = W0 + AQ B_Q ]
Only ( AQ ) and ( BQ ) are learned during fine-tuning, dramatically reducing computational requirements while maintaining performance [92].
Data Augmentation for Enhanced Generalization
Successful ensemble implementations incorporate specialized data augmentation techniques to improve model robustness:
- Fisheye Transformation: Applied with distortion coefficients sampled uniformly from -0.9 to 0.9 to emphasize central mitotic figures by enlarging central image regions [92]
- Fourier Domain Adaptation (FDA): Performs unsupervised style transfer using target images from ImageNet to enhance domain generalization, applied with probability p=0.5 [92]
- Posterization Feature Augmentation: Applied to feature crops after flipping augmentations to improve robustness to staining variations [8]
Ablation studies demonstrate that fisheye augmentation alone improves balanced accuracy by 2.3% over unaugmented models, highlighting its importance for mitosis classification [92].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents and Computational Resources
| Resource | Type/Function | Application in Ensemble Methods |
|---|---|---|
| UNI Foundation Model | Vision Transformer Architecture | Feature extraction with strong morphological pattern recognition [92] [4] |
| Virchow/Virchow2 Models | CNN-Based Architecture | Complementary feature extraction trained on diverse histopathology images [92] [4] |
| CONCH Model | Vision-Language Transformer | Cross-modal alignment of images and text for enhanced representation [8] [4] |
| Low-Rank Adaptation (LoRA) | Parameter-Efficient Fine-Tuning | Adapts foundation models to specific tasks with minimal trainable parameters [92] |
| Fisheye Transformation | Data Augmentation | Emphasizes central image regions for fine-grained cellular classification [92] |
| Fourier Domain Adaptation | Domain Adaptation | Reduces domain shift through style transfer [92] |
| Multiple Instance Learning | Weakly Supervised Learning | Aggregates patch-level predictions to slide-level diagnoses [4] |
Ensemble strategies represent a paradigm shift in computational pathology, moving beyond reliance on single models to integrated systems that leverage the complementary strengths of multiple foundation models. The experimental evidence demonstrates that ensembles consistently outperform individual models across diverse diagnostic tasks, with performance improvements of approximately 5% in balanced accuracy for challenging applications like atypical mitosis classification [92]. The key to successful implementation lies in selecting models with diverse architectural backgrounds, employing weighted fusion strategies that optimize for balanced accuracy, and incorporating uncertainty quantification to enhance reliability. As foundation models continue to evolve in scale and sophistication, ensemble methodologies will play an increasingly vital role in translating their capabilities into clinically impactful tools for precise diagnosis and personalized treatment planning.
The integration of artificial intelligence (AI) into healthcare demands robust validation frameworks to ensure tools are safe, effective, and clinically meaningful. Within computational pathology, where AI models interpret complex whole-slide images (WSIs) to aid diagnosis and prognosis, establishing trust is paramount [1]. Validation provides the foundational evidence that these tools perform as intended in their specific context of use. This guide explores the core validation frameworks essential for regulatory approval and clinical adoption, with a specific focus on the emerging paradigm of pathology foundation models compared to traditional transfer learning approaches.
The V3 framework—encompassing Verification, Analytical Validation, and Clinical Validation—has become a cornerstone for evaluating digital health technologies [94]. Originally developed for clinical Biometric Monitoring Technologies (BioMeTs), this framework is being adapted for preclinical and nonclinical contexts, including in vivo digital measures, highlighting its versatility [95]. In parallel, regulatory bodies provide guidance on Analytical Validity, Clinical Validity, and Clinical Utility for In Vitro Diagnostics (IVDs), creating a parallel structure for assessing diagnostic tools [96]. Understanding these frameworks is the first step in translating a promising algorithm into a clinically deployed tool.
Core Clinical Validation Frameworks
The V3 Framework: Verification, Analytical Validation, and Clinical Validation
The V3 framework offers a structured, three-stage approach to build a body of evidence for digital medicine products [94].
- Verification answers the question: "Was the system built correctly?" It is a systematic evaluation, often at the bench, to ensure the hardware and software components of a tool correctly capture and process raw data without errors. In computational pathology, this involves confirming that the digital slide scanner and image preprocessing steps function as specified [95] [94].
- Analytical Validation answers the question: "Does the tool measure what it claims to measure?" This step assesses the performance of the algorithm itself. It evaluates the precision and accuracy of the algorithm in transforming its input (e.g., a whole-slide image) into a defined output (e.g., a cancer detection score) [95] [94]. For a cancer grading model, analytical validation would involve establishing its accuracy in identifying and classifying tumor grades against a pathologist-generated ground truth.
- Clinical Validation answers the question: "Does the measurement matter clinically?" This final step confirms that the tool's output accurately identifies, measures, or predicts a meaningful clinical, biological, or functional state within a defined context of use and specific patient population [95] [94]. A model might perfectly detect tumor cells (analytical validation), but its clinical validation would prove that its use leads to improved patient outcomes, such as more accurate prognostication or better treatment selection.
Table 1: Components of the V3 Framework [95] [94]
| Component | Core Question | Focus of Evaluation | Typical Setting |
|---|---|---|---|
| Verification | Was the system built correctly? | Hardware/software data capture and processing | In silico / In vitro |
| Analytical Validation | Does it measure the target accurately? | Algorithm performance and output accuracy | In vivo / Clinical samples |
| Clinical Validation | Does the measurement matter clinically? | Association with clinically meaningful endpoints | Defined patient population |
The Regulatory Framework: Analytical Validity, Clinical Validity, and Clinical Utility
Parallel to V3, a framework centered on Analytical Validity, Clinical Validity, and Clinical Utility is commonly used in the regulation of IVDs and aligns with the requirements of bodies like the US FDA [96]. The concepts are highly congruent with the V3 framework but are tailored for diagnostic tests.
- Analytical Validity is nearly synonymous with the analytical validation component of V3, assessing a test's ability to accurately and reliably measure the analyte of interest.
- Clinical Validity establishes how well the test identifies, detects, or predicts a specific clinical condition or phenotype. This closely mirrors the objective of clinical validation in the V3 framework.
- Clinical Utility moves a step further, assessing whether the use of the test in clinical practice leads to improved patient outcomes and whether the benefits outweigh any risks [96].
The following diagram illustrates the logical sequence and key questions of this integrated validation pathway.
Foundation Models vs. Traditional Transfer Learning: A Validation Perspective
The choice of underlying AI methodology—emerging foundation models or established transfer learning—profoundly influences the validation strategy, with each presenting distinct advantages and challenges.
Traditional Transfer Learning
This approach typically involves taking a model pre-trained on a large dataset of natural images (e.g., ImageNet) and fine-tuning it on a smaller, targeted set of pathology images [97]. While this helps overcome data scarcity, the significant differences in image content and statistics between natural images and histology can limit transferability. Studies have shown that the general knowledge transferred resides mainly in the early layers of the network, with deeper layers offering marginal gains at a high cost of model complexity [97]. From a validation standpoint, this often means that each new diagnostic task (e.g., breast cancer detection, lung cancer subtyping) requires building a new model and conducting a full, independent V3 process, which is resource-intensive.
Pathology Foundation Models
Foundation models represent a paradigm shift. These are large AI models pre-trained on massive, diverse datasets of histopathology images using self-supervised learning, which does not require manual annotations [8] [32]. They learn general-purpose, transferable representations of histopathological morphology. Examples include UNI, trained on over 100 million images from 100,000+ WSIs [32], and TITAN, a multimodal model trained on 335,645 WSIs aligned with pathology reports [8]. These models act as versatile feature extractors that can be adapted to many downstream tasks with minimal task-specific data.
The key advantage for validation is that the verification and analytical validation of the core model can be established once, at the foundation level. When deployed for a new clinical task, the validation effort can then focus on the clinical validation of the task-specific adapter, significantly streamlining the pathway to regulatory approval and clinical adoption for multiple applications.
Table 2: Comparison of Foundation Models and Traditional Transfer Learning in Computational Pathology
| Feature | Foundation Models | Traditional Transfer Learning |
|---|---|---|
| Pre-training Data | Massive-scale, diverse histopathology WSIs (e.g., 100k+ slides) [32] | Natural image datasets (e.g., ImageNet) [97] |
| Primary Method | Self-supervised learning on histology patches/WSIs [32] | Supervised pre-training on natural images |
| Representation | General-purpose, histology-specific features | General-purpose, natural image features |
| Key Advantage | High performance across diverse tasks; data efficiency [32] [21] | Leverages existing models; avoids training from scratch |
| Key Limitation | Massive computational and data curation resources required | Limited by domain shift from natural to pathology images [97] |
| Impact on Validation | Core model validated once; task-specific validation simplified | Requires full V3 process for each new task and model |
Performance Comparison and Experimental Data
Rigorous benchmarking studies and model evaluations provide quantitative evidence of the performance advantages offered by foundation models.
Benchmarking Performance
A comprehensive 2025 benchmarking study of 31 AI foundation models demonstrated that pathology-specific vision models (Path-VMs) delivered the highest performance across a wide range of tasks, outperforming both general vision models and pathology-specific vision-language models [21]. This underscores the value of domain-specific pre-training. Furthermore, the study found that model size and data size did not consistently correlate with performance, challenging simple scaling assumptions and highlighting the importance of data diversity and model architecture [21].
Performance Across Key Tasks
Foundation models have set new state-of-the-art benchmarks across clinically relevant tasks. For instance:
- UNI was evaluated on a challenging 108-class cancer type classification (OncoTree code) and demonstrated superior performance compared to previous state-of-the-art models like CTransPath, which was pre-trained on a smaller dataset [32]. The scaling capabilities of UNI showed that increasing the scale of pre-training data directly improved performance on complex, rare cancer classification tasks [32].
- PathOrchestra, another foundation model, was evaluated on 112 diverse tasks. It achieved an accuracy exceeding 0.950 in 47 tasks, including pan-cancer classification, lymphoma subtyping, and bladder cancer screening, demonstrating remarkable robustness and generalizability [98].
- TITAN, a multimodal foundation model, excels in zero-shot classification and cross-modal retrieval (e.g., finding similar WSIs based on a text query from a pathology report), capabilities that are absent in traditional transfer learning models [8].
Table 3: Experimental Performance of Select Pathology Foundation Models
| Model (Year) | Pre-training Data Scale | Key Reported Performance Highlights | Significance for Clinical Adoption |
|---|---|---|---|
| UNI [32] | 100,426 WSIs, 20 tissue types | Outperformed prior models (CTransPath, REMEDIS) on 108-class OncoTree cancer classification. | Demonstrates scalability and superior generalization to rare cancers, a key clinical challenge. |
| PathOrchestra [98] | 287,424 WSIs, 21 tissue types | Achieved accuracy >0.950 in 47/112 tasks, including pan-cancer classification and lymphoma subtyping. | High accuracy across a vast task portfolio indicates strong clinical readiness and versatility. |
| TITAN [8] | 335,645 WSIs | Outperformed slide and region-of-interest (ROI) models in zero-shot and few-shot learning. | Reduces reliance on large labeled datasets, enabling application in resource-limited scenarios. |
Detailed Experimental Protocols
To ensure reproducibility and provide a clear roadmap for researchers, this section outlines the core experimental methodologies used to validate foundation models.
Self-Supervised Pre-training of Foundation Models
The workflow for pre-training a foundation model like UNI or TITAN involves several standardized steps, from data curation to model optimization, as visualized below.
- Dataset Curation and Preprocessing: A large-scale dataset of H&E-stained WSIs is assembled, ensuring diversity across organ types, specimen types (FFPE/frozen), and scanners [98] [32]. Each WSI is divided into smaller, manageable image patches (e.g., 256x256 pixels at 20x magnification).
- Self-Supervised Learning (SSL): A model (typically a Vision Transformer or ConvNet) is trained on the patches without using human-provided labels. Algorithms like DINOv2 [32] or iBOT [8] are used. These methods create learning objectives by, for instance, comparing different augmented views of the same image patch, forcing the model to learn robust, general-purpose features of histology.
- Feature Embedding Generation: After pre-training, the model serves as a powerful feature extractor. Any new image patch can be passed through the model to obtain a numerical vector (an "embedding") that represents its core morphological characteristics.
Downstream Task Validation Protocol
Once a foundation model is pre-trained, its utility is tested on specific clinical tasks using the following protocol:
- Task Definition and Dataset Preparation: A downstream task is defined (e.g., breast cancer metastasis detection in lymph nodes). A separate dataset with slide-level or region-level labels is prepared, split into training, validation, and held-out test sets.
- Feature Extraction and Model Adaptation: The foundation model is used to extract feature embeddings from all patches in the downstream task's WSIs. These features are then used to train a simple task-specific classifier, such as:
- Linear Probing: A single linear layer is trained on top of the frozen features. This tests the quality of the features themselves [98].
- Attention-Based Multiple Instance Learning (ABMIL): This is a standard method for slide-level prediction. It aggregates patch-level features into a single slide-level prediction, learning to weight the importance of different patches [32].
- Performance Benchmarking: The model's performance is evaluated on the held-out test set using metrics like Area Under the Curve (AUC), Accuracy (ACC), and F1-score. Its results are compared against benchmarks, including models trained via traditional transfer learning, to establish superiority [32] [21].
The Scientist's Toolkit: Essential Research Reagents and Materials
Successfully developing and validating models in computational pathology relies on a suite of key resources and tools.
Table 4: Essential Research Reagents and Solutions for Computational Pathology Validation
| Tool / Resource | Function and Role in Validation | Examples / Specifications |
|---|---|---|
| Whole-Slide Image Scanners | Digitizes glass slides to create high-resolution WSIs; critical for Verification. | Aperio ScanScope, 3DHISTECH Pannoramic, Philips IntelliSite [98] [1] |
| Curated WSI Repositories | Provides large-scale, diverse data for pre-training and benchmarking; foundational for Analytical and Clinical Validation. | The Cancer Genome Atlas (TCGA), in-house hospital archives, CAMELYON datasets [32] [1] |
| Pathology Foundation Models | Serves as a pre-trained, off-the-shelf feature extractor; accelerates development and standardizes feature quality. | UNI, TITAN, PathOrchestra, CTransPath [8] [98] [32] |
| Multiple Instance Learning (MIL) Frameworks | Enables slide-level prediction from patch-level features; essential for Clinical Validation on real-world data. | Attention-based MIL (ABMIL) and its variants [32] [1] |
| Benchmarking Platforms and Datasets | Standardized tasks and datasets for objective performance comparison; crucial for demonstrating competitive advantage. | TCGA, CPTAC, public challenges (e.g., CAMELYON) [21] |
Conclusion
The transition from traditional transfer learning to foundation models represents a fundamental paradigm shift in computational pathology, offering superior generalization, data efficiency, and multi-modal capabilities. Evidence from large-scale benchmarks indicates that models like CONCH and Virchow2 consistently outperform previous approaches, with vision-language models demonstrating particular promise in leveraging clinical context. However, critical challenges remain in ensuring robustness against domain shift, mitigating computational burdens, and establishing standardized clinical validation frameworks. Future progress will depend on developing more biologically-grounded architectures, creating comprehensive multi-modal datasets, and establishing rigorous clinical trial evidence. Foundation models are poised to become the core infrastructure for computational pathology, ultimately enabling more accessible, standardized, and predictive oncology diagnostics while necessitating continued collaboration between AI researchers, pathologists, and regulatory bodies to fully realize their clinical potential.
References
- 1. Computational pathology in the age of artificial intelligence [https://pmc.ncbi.nlm.nih.gov/articles/PMC12451823/]
- 2. A Comprehensive Hands-on Guide to Transfer Learning ... [https://medium.com/data-science/a-comprehensive-hands-on-guide-to-transfer-learning-with-real-world-applications-in-deep-learning-212bf3b2f27a]
- 3. Evaluation of a Task-Specific Self-Supervised Learning Framework in... [https://pubmed.ncbi.nlm.nih.gov/39455029/]
- 4. Benchmarking foundation models as feature extractors for ... [https://www.nature.com/articles/s41551-025-01516-3]
- 5. Comparing Computational using... Pathology Foundation Models [https://arxiv.org/html/2509.15482v2]
- 6. Foundational Models Explained: A Deep Dive into AI [https://ubiai.tools/foundational-models-explained-a-deep-dive-into-ai-2/]
- 7. AI Foundation Models: What Are They and Why Do ... [https://www.aidoc.com/learn/blog/introduction-to-foundation-models/]
- 8. A multimodal whole-slide foundation model for pathology [https://www.nature.com/articles/s41591-025-03982-3]
- 9. State of Foundation Model Training Report 2025 [https://neptune.ai/state-of-foundation-model-training-report]
- 10. Apple's Foundation Models framework unlocks new ... [https://www.apple.com/newsroom/2025/09/apples-foundation-models-framework-unlocks-new-intelligent-app-experiences/]
- 11. Optimization of Transfer Learning of Foundation Models for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12534908/]
- 12. Towards Understanding and Harnessing the Transferability ... [https://arxiv.org/html/2508.13482v1]
- 13. Fine-tuning foundation models of materials interatomic ... [https://www.nature.com/articles/s41524-025-01727-x]
- 14. CNN vs Vision Transformer (ViT): Which Wins in 2025? [https://hiya31.medium.com/cnn-vs-vision-transformer-vit-which-wins-in-2025-e1cb2dfcb903]
- 15. Pre-trained convolutional neural network with transfer ... [https://pubmed.ncbi.nlm.nih.gov/39904596/]
- 16. Vision Transformers for Low-Quality Histopathological ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11817517/]
- 17. Transformer optimization with meta learning on pathology ... [https://www.nature.com/articles/s41746-025-01833-6]
- 18. Evaluating vision transformers and convolutional neural ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12522997/]
- 19. Histopathological classification of colorectal cancer based ... [https://www.nature.com/articles/s41598-025-19134-z]
- 20. Transfer Learning-Based Ensemble of CNNs and Vision ... [https://www.mdpi.com/2075-4418/15/15/1928]
- 21. Evaluating Vision and Pathology Foundation Models for ... [https://pubmed.ncbi.nlm.nih.gov/40463538/]
- 22. Rethinking Foundation Models in Pathology: A New Lens [https://beyondtheslide.substack.com/p/rethinking-foundation-models-pathology?utm_source=substack&utm_medium=email&utm_content=share&action=share]
- 23. Preparing Data for Artificial Intelligence in Pathology with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10572440/]
- 24. Unleashing the potential of digital pathology data ... [https://www.nature.com/articles/s41746-022-00635-4]
- 25. Comparison of whole slide imaging devices in a clinical ... [https://www.sciencedirect.com/science/article/pii/S2153353925000318]
- 26. A quantitative approach to evaluate image quality of whole ... [https://www.sciencedirect.com/science/article/pii/S2153353922005740]
- 27. Comparative analysis of supervised and self ... [https://www.nature.com/articles/s41598-025-99000-0]
- 28. Scaling Self - Supervised for Learning with Histopathology ... Masked [https://www.medrxiv.org/content/10.1101/2023.07.21.23292757v3.full]
- 29. Self supervised contrastive learning for digital histopathology [https://arxiv.org/abs/2011.13971]
- 30. Self supervised contrastive learning for digital histopathology [https://www.sciencedirect.com/science/article/pii/S2666827021000992]
- 31. Scaling Self - Supervised for Learning with Histopathology ... Masked [https://www.owkin.com/publications/scaling-self-supervised-learning-for-histopathology-with-masked-image-modeling]
- 32. Towards a General-Purpose Foundation Model for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11403354/]
- 33. AI in Histopathology Explorer for comprehensive analysis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11904230/]
- 34. Self-supervised identification and elimination of harmful ... [https://pubmed.ncbi.nlm.nih.gov/39955398/]
- 35. Optimization of Transfer Learning of Foundation Models for ... [https://pubmed.ncbi.nlm.nih.gov/41112860/]
- 36. A Systematic Review of Vision and Vision-Language ... [https://www.sciencedirect.com/science/article/pii/S2667376225000514]
- 37. Unlocking the potential: multimodal AI in biotechnology ... [https://www.nature.com/articles/s41746-025-01992-6]
- 38. From siloed data to breakthroughs: multimodal AI in drug ... [https://www.drugtargetreview.com/article/160597/from-siloed-data-to-breakthroughs-multimodal-ai-in-drug-discovery/]
- 39. Foundation Models for Computational Pathology [https://www.sciencedirect.com/special-issue/324717/foundation-models-for-computational-pathology]
- 40. A visual-language foundation model for computational ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11384335/]
- 41. Virchow 2: Scaling Self-Supervised Mixed Magnification ... [https://arxiv.org/html/2408.00738v1]
- 42. mahmoodlab/CONCH: Vision-Language Pathology ... [https://github.com/mahmoodlab/CONCH]
- 43. A foundation model for clinical-grade computational ... [https://www.nature.com/articles/s41591-024-03141-0]
- 44. Boost foundation model results with linear probing and fine ... [https://snorkel.ai/blog/boost-foundation-model-results-with-linear-probing-fine-tuning/]
- 45. Evaluation of Few-Shot Learning for Classification Tasks in ... [https://arxiv.org/abs/2404.17832]
- 46. Fully fine-tuned CLIP models are efficient few-shot learners [https://www.sciencedirect.com/science/article/abs/pii/S0950705125008652]
- 47. CLAP: Enhancing Linear Probing for Efficient Few-Shot ... [https://datascienceharp.medium.com/clap-enhancing-linear-probing-for-efficient-few-shot-learning-in-vision-language-models-2681c01a699d]
- 48. Scaling beyond the Limit of Few-shot In-Context Learners [https://arxiv.org/abs/2212.10873]
- 49. PMI estimation with cross-species transfer and visual... learning [https://link.springer.com/article/10.1007/s00414-025-03659-z]
- 50. Evaluation of Few-Shot Learning for Classification Tasks ... [https://ui.adsabs.harvard.edu/abs/2024arXiv240417832H/abstract]
- 51. GitHub - mahmoodlab/UNI: Pathology - Nature... Foundation Model [https://github.com/mahmoodlab/UNI]
- 52. Deep Learning-Based Prediction of Molecular Tumor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9784641/]
- 53. Real-world deployment of a fine-tuned pathology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12443599/]
- 54. PLUTO-4: Frontier Pathology Foundation Models [https://arxiv.org/html/2511.02826v3]
- 55. Artificial intelligence-based digital pathology using H&E ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12323524/]
- 56. Development and clinical validation of deep learning-based ... [https://bmcgastroenterol.biomedcentral.com/articles/10.1186/s12876-025-04045-0]
- 57. Computational pathology: A comprehensive review of ... [https://www.sciencedirect.com/science/article/pii/S2950261625000214]
- 58. Advancements in Radiology Report Generation [https://pmc.ncbi.nlm.nih.gov/articles/PMC12292164/]
- 59. 2025 Expert Consensus on Retrospective Evaluation of ... [https://www.sciencedirect.com/science/article/pii/S2667102625001044]
- 60. The Rise of AI-Assisted Diagnosis: Will Pathologists Be ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12468387/]
- 61. Investigation on potential bias factors in histopathology ... [https://www.nature.com/articles/s41598-025-89210-x]
- 62. Learning generalizable AI models for multi-center ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11271637/]
- 63. Domain Shift in Prostate MRI AI: Equipment & Protocols [https://botimageai.com/prostate-mri-ai-domain-shift-equipment-protocols/]
- 64. Unveiling Institution-Specific Bias in Pathology Foundation ... [https://arxiv.org/html/2502.16889v1]
- 65. A clinical benchmark of public self-supervised pathology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12003829/]
- 66. AdverIN: Monotonic Adversarial Intensity Attack for Domain ... [https://www.sciencedirect.com/science/article/abs/pii/S1361841525003949]
- 67. Multicenter validation of a scalable, interpretable, multitask ... [https://www.nature.com/articles/s41746-025-01949-9]
- 68. Domain-Shift-Aware Conformal Prediction for Large ... [https://arxiv.org/html/2510.05566v1]
- 69. Do Multiple Instance Learning Models Transfer? [https://arxiv.org/html/2506.09022v1]
- 70. Foundation model [https://en.wikipedia.org/wiki/Foundation_model]
- 71. Deep learning with transfer learning in pathology. Case study [https://pmc.ncbi.nlm.nih.gov/articles/PMC9289702/]
- 72. Do Multiple Instance Learning Models Transfer? [https://arxiv.org/abs/2506.09022]
- 73. AI-augmented pathology: the experience of transfer ... [https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2025.1598289/full]
- 74. Adversarial attacks and adversarial robustness in ... [https://www.nature.com/articles/s41467-022-33266-0]
- 75. Why Foundation Models in Pathology Are Failing [https://arxiv.org/html/2510.23807v1]
- 76. A whole-slide foundation model for digital pathology from ... [https://www.nature.com/articles/s41586-024-07441-w]
- 77. Comparing Computational Pathology Foundation Models ... [https://arxiv.org/abs/2509.15482]
- 78. Closing the gap in the clinical adoption of computational ... [https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-025-01484-y]
- 79. integrating explainable AI (XAI) with machine learning ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12465982/]
- 80. Medical artificial intelligence and the black box problem [https://www.sciencedirect.com/science/article/pii/S2667102623000578]
- 81. The “Black Box” Problem in Healthcare AI: Building Trust ... [https://medium.com/@healthai_official/the-black-box-problem-in-healthcare-ai-building-trust-and-transparency-4b8004b4a08b]
- 82. 11 ML Performance Benchmarking Tools You Should Know [https://overcast.blog/11-ml-performance-benchmarking-tools-you-should-know-20ad8eee9e3a]
- 83. A Survey on Computational Pathology Foundation Models [https://arxiv.org/html/2501.15724v2]
- 84. How to Train a State-of-the-Art Pathology Foundation ... [https://sophontai.com/blog/openmidnight]
- 85. Evaluating Vision and Pathology Foundation Models for ... [https://www.medrxiv.org/content/10.1101/2025.05.08.25327250v1.full-text]
- 86. Why Foundation Models in Pathology Are Failing [https://arxiv.org/html/2510.23807v2]
- 87. Computational pathology in precision oncology: From task- ... [https://www.eurekalert.org/news-releases/1105085]
- 88. Vision Language Models (Better, faster, stronger) [https://huggingface.co/blog/vlms-2025]
- 89. PathVLM-Eval: Evaluation of open vision language models ... [https://www.sciencedirect.com/science/article/pii/S2153353925000409]
- 90. Investigating Zero-Shot Diagnostic Pathology in Vision ... [https://arxiv.org/abs/2505.00134]
- 91. A Survey of Vision-Language Interactive Reasoning in ... [https://arxiv.org/html/2509.25373v1]
- 92. Ensemble of Pathology Foundation Models for MIDOG ... [https://arxiv.org/html/2509.02591v1]
- 93. Uncertainty-aware ensemble of foundation models ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12480093/]
- 94. Verification, analytical validation, and clinical validation (V3) [https://www.nature.com/articles/s41746-020-0260-4]
- 95. Validation framework for in vivo digital measures - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11751004/]
- 96. Framework for Developing Clinical Evidence ... [https://mdic.org/resources/framework-for-developing-clinical-evidence-for-regulatory-and-coverage-assessments-in-in-vitro-diagnostics/]
- 97. How much off-the-shelf knowledge is transferable from natural images... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7556818/]
- 98. PathOrchestra: a comprehensive foundation model for ... [https://www.nature.com/articles/s41746-025-02027-w]